
□ RNASTAR: An RNA STructural Alignment Repository that provides insight into the evolution of natural and artificial RNAs
>> http://rnajournal.cshlp.org/content/18/7/1319.full
RNA構造アライメント・リポジトリ: 単結晶またはNMR構造にバックアップされた、9600のユニーク解析を手動で配列したキュレーション。自動化アライメントの改善に応用。

□ geneXplain:
In silico method for modelling metabolism and gene product expression at genome scale : Nature Communications http://bit.ly/NasH3w
in silico comparative transcriptomics, the model allows the discovery of new regulons and improving the genome and transcription unit annotations. Our method presents a framework for investigating molecular biology and cellular physiology in silico and may allow quantitative interpretation of multi-omics data sets in the context of an integrated biochemical description of an organism.

□ Technical and biological variance structure in mRNA-Seq data: life in the real world
>> http://www.biomedcentral.com/content/pdf/1471-2164-13-304.pdf
In mRNA-Seq data from 25 subjects, we found technical variation to generally follow a Poisson distribution as has been reported previously and biological variability was over- dispersed relative to the Poisson model. The mean-variance relationship across all genes was quadratic, in keeping with a Negative Binomial (NB) distribution.
Over-dispersed Poisson and NB distributional assumptions demonstrated marked improvements in goodness-of-fit (GOF) over the standard Poisson model assumptions, but with evidence of over-fitting in some genes. Modeling of experimental effects improved GOF for high variance genes but increased the over-fitting problem.
mRNAの配列データと生物学的分散構造: 過分散ポアソン、NB分布からカウントデータを改善。生体信号とデータの意味
□ m_m_campbell:
Mind the gap. New algorithms correct DNA sequencing errors, reducing gaps. http://bit.ly/ObEBi8 #genomics #bioinformatics
Correction algorithms extend the reach of genome sequencing:
‘Second-generation sequencers’ read genomes in pieces of 100?700 base pairs long, but those pieces are hard to stitch together in the correct order. ‘Third-generation’, or single-molecule sequencers such as the PacBio RS, made by Pacific Biosciences, produce reads as long as 23,000 bases, but make more errors than typical genome-analysis software can tolerate.
Reserchers used short reads from second-generation Illumina or Roche 454 machines to correct errors in long single-molecule reads from the PacBio RS. They tested their correction algorithm on the genomes of Escherichia coli and yeast, as well as the collection of messenger RNA, or transcriptome, of maize, and found they could improve accuracy, from roughly 83% to as high as 99.9%. They also applied this hybrid correction strategy to a previously unsequenced genome of a parrot (Melopsittacus undulates).
補正アルゴリズムを用い、2nd - 3rd-generation シーケンサーを併用して解析
□ The Return of Finished Genomes: Hybrid Sequencing Strategy Boosts Pacific Biosciences Accuracy, Assembly http://www.bio-itworld.com/news/07/02/12/Return-finished-genomes-hybrid-sequencing-boosts-PacBio-accuracy-assembly.html
□ hinaichigo:
PacBio ALLORA assemblerか。日本語ブログでいち早く紹介してるのがショートリードの憂鬱の方か。http://pacbiobrothers.blogspot.jp/ ALLORAが外部持って行けるなら、sangerで組んだミドルサイズcontigと合わせる技に応用できるかも
□ A systematic comparison and evaluation of high density exon arrays and RNA-seq technology used to unravel the peripheral blood transcriptome of sickle cell disease. http://www.ncbi.nlm.nih.gov/pubmed/22747986
Affymetrix Exonアレイとイルミナシーケンサの体系的比較と使い分け
解析パイプラインやモジュールはともかく、2nd-3rdシーケンサ単位で併用する場合、プラットフォームのプロトコルは規格を合議した方が良いのではないかと思うのだけど、似た理由で直近のメーカーやサプライヤ間の業務提携が相次いでるのだし、サービスプロバイダに担保するより一般化したら良い。
□ CSHLnews:
How error-corrected 3rd-gen #sequencing gave unprecedented glimpse of speech-gene homologs in parrot: http://bit.ly/NCNxZh #biotechnology
□ n0rr:
シーケンサーの進歩の影響度合い:データの質>データ解析の技術>データ解釈する技術>サンプル作る技術 左の方について行けないなら(中米に置いて行かれるなら)右の方で勝負すれば良い。ひとつ心配なのが実験デザインはどこにくるんだろうってところ。

□ Quantifying and Analyzing the Network Basis of Genetic Complexity:
>> http://t.co/bEjRJeNF
The definition equates genetic complexity with a surplus of genotypic diversity over phenotypic diversity. Applying this definition to ensembles of Boolean network models, we found that the in-degree distribution and the number of periodic attractors produced determine the relative complexity of different topology classes.
The rigorous definition of genetic complexity is a tool for unraveling the structure and properties of genotype-to-phenotype maps by enabling the quantitative comparison of the relative complexities of different genetic systems.
遺伝子型-表現型マップの複雑性をブーリアンネットワークのトポロジカルモチーフで定量化・分析する。
"Conserved topological motifs impart similar properties in different biological systems” 遺伝子ネットワークのトポロジーは生物学的システムに類似する。 この視点に目からウロコ。
□ Linux Kernel Development Visualization (git commit history - past 6 weeks - june 02 2012)
>> http://code.google.com/p/gource/
□ Transcriptional interference networks coordinate the expression of functionally related genes clustered in the same genomic loci
>> http://www.frontiersin.org/Non-Coding_RNA/10.3389/fgene.2012.00122/full
The TIN represents an auto-regulatory system with an exquisitely timed and highly synchronized cascade of gene expression in functionally linked genes located in close physical proximity to each other. In this study, we focused on herpesviruses. The reason for this lies in the compressed nature of viral genes, which allows a tight regulation and an easier investigation of the transcriptional interactions between genes.
『Transcriptional interference networks (転写干渉ネットワーク): The TIN hypothesis: TIN仮説は、 Antisense RNAやウィルス、哺乳動物細胞に至まで、あらゆる生物学的ゲノムにおけるタンデム遺伝子群の発現を説明する試みで提唱された』
TIN仮説では、遺伝子座に近接する機能関連遺伝子への高同期カスケードな自動規制システムを想定。滝モデルはタンデム配列のクラスタ内の上流遺伝子と下流遺伝子の転写の単方向性を説明。真核生物の転写調節、哺乳類Hox遺伝子群など様々な分類群でアンチセンスRNA発現の概念的枠組みを提供する.

□ Modeling Boundary Vector Cell Firing Given Optic Flow as a Cue:
>> http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1002553
The modeling work presented here investigates the role of optic flow, the apparent change of patterns of light on the retina, as input for boundary vector cell firing. Analytical spherical flow is used by a template model to segment walls from the ground, to estimate self-motion and the distance and allocentric direction of walls, and to detect drop-offs.
Distance estimates of walls in an empty circular or rectangular box have a mean error of less than or equal to two centimeters. Integrating these estimates into a visually driven boundary vector cell model leads to the firing patterns characteristic for boundary vector cells. This suggests that optic flow can influence the firing of boundary vector cells.
モデリング境界ベクター細胞の光フロー発火。網膜上の光パターン球状フローと自己運動パラメータのテンプレートモデル

□ Sagace: Search for Biomedical Data & Resources in Japan: Informative search results with metadata (taxonomyID, DB name) http://sagace.nibio.go.jp/en/index.html
□ Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses http://pubs.rsc.org/en/Content/ArticleLanding/2012/CS/c1cs15258k
Polymeric materials have been extensively developed as a delivery vehicle for nucleic acids over the past two decades. Many previous studies have demonstrated that synthetic delivery vehicles can be highly functionalized by chemical approaches to overcome biological barriers in nucleic acid delivery, similar to viruses.
Based on our current knowledge, this tutorial review describes rational strategies in the design of polymeric materials to achieve construction of the versatile vehicles, that is “artificial viruses”, for successful gene therapy, especially focusing on the chemical structures with the minimal adverse effects.
□ Mount Sinai Team Reports Data Showing Most miRNAs Have Low Suppressive Activity
>> http://t.co/cMCcGdxb
□ genetics_blog:
bitseq - Bayesian Inference of Transcripts from Sequencing Data for RNA-seq http://code.google.com/p/bitseq/wiki/BitSeq #bioinformatics

□ Lists of Genomics Software/Service Providers:
世界のゲノム、Bioinformatics関連企業、プロバイダ 一覧。実績企業をまとめてるので、これから市場調査したい人には便利かも。 http://grouthbio.com/Genome_Software_Service.php

□ Computational Methods for de novo Assembly of Next-Generation Genome Sequencing Data
>> http://perso.eleves.bretagne.ens-cachan.fr/%7Echikhi/RChikhi-Thesis-Slides.pdf
Definition of the assembly problem Contributions
Contribution 1 : localized assembly Index
Contribution 2 : incorporation of pairing information Monument assembler
Contribution 3 : ultra-low memory assembly Minia
次世代シーケシングにおけるアセンブリの問題を、大きく三つに定義。ブルームフィルタに基づくde BrujinのMinia法が凄い。
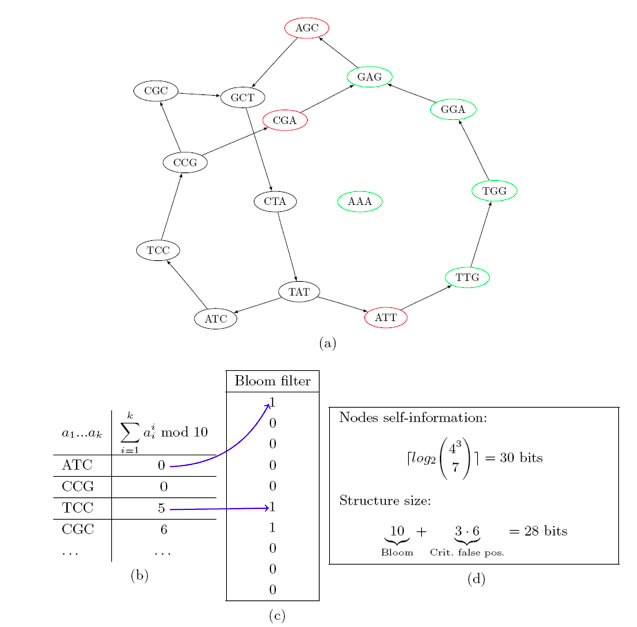
□ Minia: Space-efficient & exact de Bruijn graph representation based on a Bloom filter:
ヒトゲノムNA18507のリードに対し23時間5.7 GBで読み込み
>> http://minia.genouest.org/minia.pdf
□ Drug & Diagnostic Development 2012:
□ Next-Gen Sequencing Applications & Translational Technologies Aug6-8, 2012, SF, CA
次世代シーケンシングの応用と翻訳のための技術-トランスレーショナル生物学
>> http://www.ibclifesciences.com/Sequencing/overview.xml
>> http://www.gii.co.jp/conference/ibu223128-2012/sequencing.shtml
・NGS to make therapeutic decisions
・Clinical applications in oncology
・Discovery and clinical trial applications
・Third-generation sequencing
・NGS applied for clinical diagnostics
・Technologies to reveal disease biology
・NGS in carrier screening and prenatal Dx
・CTCs, non-coding RNAs, single cell systems
□ AgilentLife:
Agilent Names IMAXIO as Certified Provider of Target Enrichment Services for Next-Gen Sequencing - http://bit.ly/P32lX9
□ Molecular Identification of Commercialized Medicinal Plants in Southern Morocco: モロッコの薬用植物インベントリにDNAバーコーディングを実施。漢方薬に続き。 http://www.plosone.org/article/info:doi/10.1371/journal.pone.0039459
「ITSの参照配列からは34%の多型性、及び11%の真菌汚染が見られた」市場標本と参照種の同定は困難そう。リファレンス・データベースはGenBankに対しMegablastアルゴリズムを使用して識別。Blastclust分析により、クエリシーケンスを基準配列に基づいて同定。
□ MuSiC: Identifying mutational significance in cancer genomes: 癌ゲノムの包括的変異パイプライン。大規模コホートにおけるシーケンスベースの入力を標準化し、真に重要な事象を分離する http://genome.cshlp.org/content/early/2012/07/02/gr.134635.111.abstract
Mutational Significance in Cancer (MuSiC)
comprehensive mutational analysis pipeline that uses standardized sequence-based inputs along with multiple types of clinical data to establish correlations among mutation sites, affected genes and pathways, and to ultimately separate the commonly abundant passenger mutations from the truly significant events.
□ rnomics:
Chimeras taking shape: Potential functions of proteins encoded by chimeric #RNA transcripts [RESEARCH]:... http://bit.ly/MNOGyC
□ rnomics:
Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between #mRNA synthesis and... http://bit.ly/MNOEqh
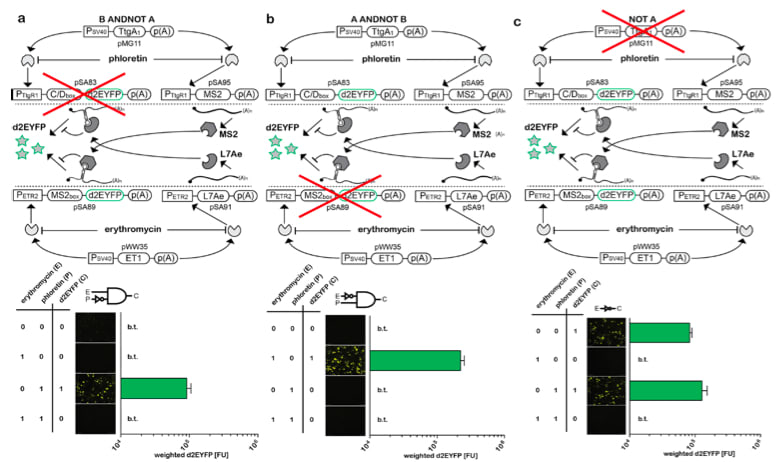
□ Programmable single-cell mammalian biocomputers
>> http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11149.html
Rational interconnection of these synthetic switches resulted in increasingly complex designer networks that execute input-triggered genetic instructions with precision, robustness and computational logic reminiscent of electronic circuits2, 3. Using trigger-controlled…
プログラマブルな哺乳類由来の単一細胞を用いた生命計算回路: 生物のプログラム細胞と遺伝的相互作用を用いて、高精度かつ堅牢な計算ロジックを構築 。
・Supplementary Information
>> http://www.nature.com/nature/journal/vaop/ncurrent/extref/nature11149-s1.pdf
この下段にフローサイトメトリー顕微鏡下の回路エンコーディングプラスミドのトランスフェクション量と細胞応答の相関。哺乳類細胞バイオ計算の臨床応用に鍵
□ CPSS: a computational platform for the analysis of small RNA deep sequencing data: http://mcg.ustc.edu.cn/db/cpss/index.html
□ Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening http://www.nature.com/nsmb/journal/vaop/ncurrent/full/nsmb.2334.html
□ Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease
>> http://www.nature.com/nrn/journal/v13/n7/full/nrn3271.html
□ RIP-chip-SRM?a new combinatorial large-scale approach identifies a set of translationally regulated bantam/miR-58 http://genome.cshlp.org/content/22/7/1360.full?rss=1&utm_source=dlvr.it&utm_medium=twitter
□ Triplexator: Detecting nucleic acid triple helices in genomic & transcriptomic data: ゲノム・トランスクリプトームデータからの三重鎖構造検出フレームワーク http://genome.cshlp.org/content/22/7/1372.abstract
Triplexator, the first computational framework that integrates all aspects of triplex formation, and showcase its potential by discussing research examples for which the different aspects of triplex formation are important.

□ NGS & RNA-Seq Form Dynamic Duo
>> http://t.co/CsVriDgt
A multilayer and integrative analysis of the whole transcriptome in gastric cancer: Tumor and noncancerous samples were first subjected to two complementary sequencing protocols that target the RNA fragments from 50?150 nucleotides and 18?40 nucleotides, respectively. Then a multilayer analysis was performed to identify different types of transcriptional aberrations that were associated with gastric cancer, including differentially expressed mRNAs, key differentially expressed miRNAs, and recurrent somatic mutation candidates. Finally, the integrative analysis suggests AMPKa2 as a potential functional target in Asian gastric cancer.
□ dgmacarthur:
If these sensible standards were applied universally, bulk of cell biology papers would evaporate: http://labstats.net/articles/cell_culture_n.html

(Multi-dimensional phenotyping)
□ Screens, maps & networks: from genome sequences to personalized medicine.
>> http://t.co/bDFhbjpI
Recently, information about the genetic makeup of cancer cells has been combined with novel functional genomics approaches to identify novel targets, exploit synthetic lethality and explore the rewiring of cellular pathways. Here, we highlight recent developments revealing the hidden landscape of genetic interactions in model organisms and cancer cells, a key step toward personalized cancer diagnostics and therapy.
□ BioLOD:
GenoCon2 Challenge B: design a DNA sequence conferring to the model organism Arabidopsis thaliana functionality... http://fb.me/1DQOk58i4
GenoCon2(合理的ゲノム設計コンテスト)のChallengeBは、前回のリベンジか。シロイヌナズナのカルビン回路に空中ホルムアルデヒド解毒機能を付与するDNA配列の設計。シーケンス機能向上に寄る改善だけでなく、全く新しいメソッドも可。
□ Dynamic kinetic energy potential for orbital-free density functional theory
>> http://jcp.aip.org/resource/1/jcpsa6/v134/i14/p144101_s1
A dynamic kinetic energy potential (DKEP) is developed for time-dependent orbital-free (TDOF) density function theory applications.
The DKEP potential should be a powerful tool for embedding a dynamical system described by a more accurate method (such as time-dependent density functional theory, TDDFT) in a large background described by TDOF with a DKEP potential.
□ New Stanford method enables sequencing of fetal genomes using only maternal blood sample: 母体の血液サンプルから胎児のゲノム・シーケシングを可能に http://www.eurekalert.org/pub_releases/2012-07/sumc-nsm070212.php
□ Ultrafast genome-wide scan for SNP-SNP interactions in common complex disease http://genome.cshlp.org/content/early/2012/07/05/gr.137885.112.abstract
□ MCM8- and MCM9-Deficient Mice Reveal Gametogenesis Defects and Genome Instability Due to Impaired Homologous Recombination http://www.cell.com/molecular-cell/abstract/S1097-2765(12)00494-7
□ Study Design Calculator for RNA-Seq: RNA-Seq effect size as the ratio of fold change to sample variance, estimate power http://voila.prognosysbio.com/static/VoilaStudyDesignPower.pdf
□ TriLinkBioTech:
A COMPLETE GENOME IN TIME displays the entire human haploid genome sequence in the dimension of time. http://ow.ly/bXAGM
□ QIAGENscience:
Recent study shows DNA methylation linked to memory loss http://bit.ly/M888Hc
□ Oxford Index: A free discovery service, across Oxford's digital academic content:
>> http://oxfordindex.oup.com/
オックスフォード大学出版局のオンライン学術リソース横断検索。
□ kaythaney:
Data from our recent survey "The future of UK's digital infrastructure" now available on @figshare. Help me sift. http://shar.es/sLtBm #ELC
□ InsideScience:
Joe Incandela/CMS presents group's combined data for mass of new particle at end of talk: 125.3+/- 0.6 GeV at 4.9 standard deviations.
combined significance of ZZ and gamma gamma is 5 sigma.
有意性5シグマはヒッグス粒子発見の最低水準に達したということ。
□ FermilabToday:
Tevatron #Higgs results. http://www.fnal.gov/pub/today/archive_2012/today12-07-02.html
□ sc_k:
Tevatron results indicate that the Higgs, if it exists, has a mass between 115 and 135 GeV/c2, or about 130 times the mass of the proton.
Tevatron saw Higgs signal in the combined CDF and DZero data in the bottom-quark decay mode with a statistical significance of 2.9 sigma.
Higgs Boson包囲網が完成しつつある模様。フェルミのTevatronでも2.9sigmaでシグナル検出とのこと。ってゆーか、以前からHiggsが想定より軽い場合は、LHCよりTevatronが捕捉に向いてるのではないかって声があったよね。
□ LouWoodley:
Kind of crossing the streams here at #lnlm12 - hearing about structural genomics and simultaneously following the Higgs tweets :)
つくづく、昔の『発見』は、新大陸だったり新種だったり、「絵の具で何が書けるか」みたいなテーマだったけれど、近現代はヒッグス粒子なり分子生物学なり、「この絵は何で書かれているのか」という方向へシフトしてるんだなー。
□ shioriH:
メタ個体群、メタ群集、メタ生態系があるのならシンク個体群ソース個体群、シンク群集ソース群集、シンク生態系ソース生態系があるはずや
□ envmic:
最近の微生物生態学系の学術誌を読んで気になる言葉。「微生物生態系」。なんじゃ,そら?湖沼生態系や森林生態系ならわかる。特定の空間を対象にしているから。生態系が対象としているのは人が任意に決めた空間。微生物は空間ではない。「生態系」の言葉に無頓着だから起こる現象。
思い返してみると、微生物相をスケーリング対象とした環境相互作用の観測の文脈で、『微生物生態系』という表現が用いられてることが多い印象。下厳密には『微生物群の任意空間における時間的ダイナミクスの生態系』というべきなのかなw
そのセンシティブさを『Ecosytem』に含意するのかもしれない
□ ふと思ったのだけど、地域毎の癌死亡率と癌罹患率を評価する場合、癌と診断されてからのステージ別の統計と経過時間までを併せたデータって取られてるのかな?

□ weskandar:
Here's the long awaited map of military men in civilian jobs .. http://el3askarmap.kazeboon.com/index.php @gottschau





















※コメント投稿者のブログIDはブログ作成者のみに通知されます