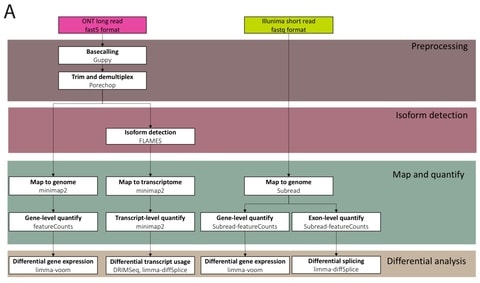
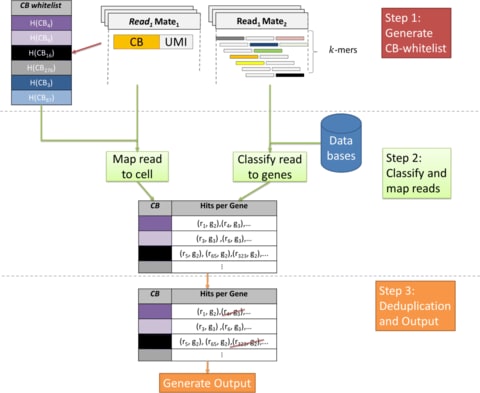
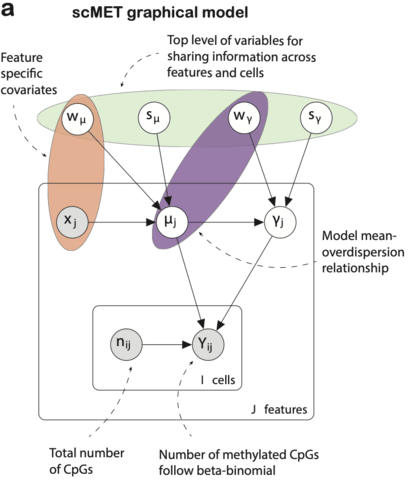
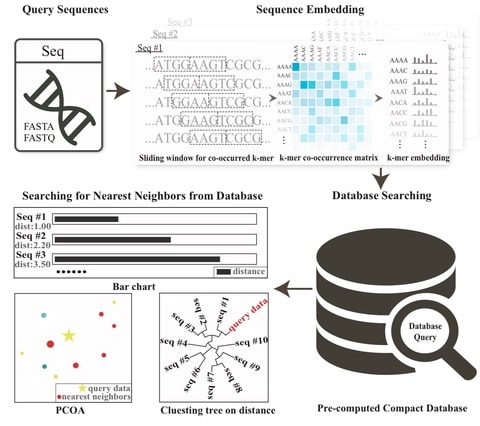
The biological knowledge is used to define only meaningful connections, shaping the architecture of the neural network. Interpretability is inherent to the neural network’s architecture.
□ IA: Efficient implied alignment
>>
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03595-2
The reduction in the time complexity of the algorithm dramatically improves both its utility in generating multiple sequence alignments and its heuristic utility.
The improvement of the IA algorithm is that the additional stored information allows us to determine the final assignments in Θ(k∗m∗n) instead of (𝑘2∗𝑛2) time. The IA algorithm can be improved to run with (𝑘∗𝑛2) and best case Ω(k∗n) complexity of time and space.

□ Pseudocell Tracer: Inferring cellular trajectories from scRNA-seq https://www.biorxiv.org/content/10.1101/2020.06.26.173179v1.full.pdf
Pseudocell Tracer uses a supervised encoder, trained with adjacent biological information, to project scRNA-seq data into a low-dimensional cellular state space.
Pseudocells are subjected to a decoder to observe gene expression dynamics along the trajectory and provide novel insights into the underlying regulatory mechanisms. Pseudocell Tracer infers trajectories in “pseudospace” rather than in “pseudotime”.

□ Pseudo-Location: A novel predictor for predicting pseudo-temporal gene expression patterns using spatial functional regression
>>
https://www.biorxiv.org/content/10.1101/2020.06.11.145565v1.full.pdf
a trajectory inference analysis in order to identify the pseudo-temporal gene expression patterns (PTGEPs) for scRNA-seq data.
pseudo-location, a new concept of genetic spatial information by incorporating the chromosome number and molecular starting position of genes. In here PTGEPs are treated as functional responses and the genetic spatial information is treated as scalar predictor.
□ Approximation of Indel Evolution by Differential Calculus of Finite State Automata
>>
https://www.biorxiv.org/content/10.1101/2020.06.29.178764v1.full.pdf
a systematic differential calculus for finding HMM-based approxmate solutions of continuous-time Markov processes on strings which are “local” in the sense that the infinitesimal generator is an HMM.
This is a reference implementation of the method to calculate alignment gap probabilities by trajectory enumeration. on the multi-residue indel process, the generality of the infinitesimal automata suggests that other local evolutionary models.

□ Compression of quantification uncertainty for scRNA-seq counts
>>
https://www.biorxiv.org/content/10.1101/2020.07.06.189639v1.full.pdf
“Pseudo-inferential” replicates were generated from a negative binomial distribution using distributional parameter values derived from the compressed uncertainty estimates.
Lineages and pseudotimes were fit using the slingshot method, and trade-Seq was used to fit the GAMs to expression counts utilizing these lineages and pseudotimes.
evaluating the impact of accounting for quantification uncertainty into trajectory-based scRNA-seq differential expression analysis using tradeSeq, and demonstrate that improvements in the false discovery rate can be obtained by incorporating pseudo-inferential replicates.
□ RNAxplorer: Harnessing the Power of Guiding Potentials for Sampling of RNA Landscapes
>>
https://www.biorxiv.org/content/10.1101/2020.07.03.186882v1.full.pdf
Most of the measures show that RNAxplorer produces more diverse structure samples and is better at finding the most relevant kinetic traps in the landscape.
RNAxplorer employs efficient dynamic programming based Boltzmann sampling, but is improved by adding guiding potentials. These potentials are accumulated into pseudo-energy terms that effectively steer sampling towards unexplored regions of the structure space.
□ ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data
>>
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03585-4
ATLAS uses additional BBTools utilities to perform an efficient error correction based on k-mer coverage (Tadpole) and paired-end read merging (bbmerge).
ATLAS uses metaSPAdes or MEGAHIT for de novo assembly, with the ability to control parameters such as k-mer lengths and k-mer step size for each assembler, as well as hybrid-assembly of paired short-and long-read libraries.

□ MRPV: Ensemble Classification through Random Projections for single-cell RNA-seq data
>>
https://www.biorxiv.org/content/10.1101/2020.06.24.169136v1.full.pdf
MRPV classification scheme, has the true potential to be established as the new default in dealing with biomedical tasks with similar characteristics. MRPV is an ensemble classification utilizing multiple ultra-low dimensional Random Projected spaces.
The MRPV approach belongs to the “parallel ensemble methods” category for which the base classifiers are constructed in parallel exploiting independence. a computationally fast, simple, yet effective approach for single cell RNA-seq data with ultra-high dimensionality.
MRPV do not require any level of approximation of the pairwise distances in the projected space, thus the resulting dimensionality r is no longer bounded by O(log n/ε2), while R does not need to be orthonormal.

□ PIDC: Gene Regulatory Network Inference from Single-Cell Data Using Multivariate Information Measures
>>
https://www.cell.com/cell-systems/fulltext/S2405-4712(17)30386-1
the methods chosen to discretize data and estimate entropies and probability distributions affect algorithm performance considerably — too often, the impact of these choices has been ignored.
PIDC, a fast, efficient algorithm that uses partial information decomposition to identify regulatory relationships between genes. PIDC allows it to outperform pairwise mutual information-based algorithms when recovering true relationships, and infer causality and directionality.

□ Fourier-transform-based attribution priors improve the interpretability and stability of deep learning models for genomics
>>
https://www.biorxiv.org/content/10.1101/2020.06.11.147272v1.full.pdf
a novel attribution prior, where the Fourier transform of input-level attribution scores are computed at training-time, and high-frequency components of the Fourier spectrum are penalized.
The prior is agnostic to the model architecture or predicted experimental assay, yet provides similar gains across all experiments. This work represents an important advancement in improving the reliability of deep learning models for deciphering the regulatory code of the genome.
□ Sparse reduced-rank regression for integrating omics data https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03606-2
a multivariate linear regression model that relates multiple predictors with multiple responses, and to identify multiple relevant predictors that are simultaneously associated with the responses.
Group Dantzig type formulation, a new computationally efficient convex formulation to estimate the coefficient matrix in that takes advantage of the potential presence of low-rankness and sparsity.

□ Circuits with broken fibration symmetries perform core logic computations in biological networks
>>
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007776
a theoretically principled strategy to search for computational building blocks in biological networks, and present a systematic route to design synthetic biological circuits.
the biological hierarchy can be extended to any number m of loops of length d and autoregulators in the fiber n, to form ever more sophisticated circuits whose complexity is expressed in generalized Fibonacci sequences Qt = nQt−1 + mQt−d.
□ GRISLI: Gene regulation inference from single-cell RNA-seq data with linear differential equations and velocity inference
>>
https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btaa576/5858974
Solving a convex regression problemno restrictive assumption on the GRN structure. These benefits come at the cost of estimating the velocity of each cell, a novel procedure based on weighted averages of finite differences with other cells at nearby positions in space-time.
GRISLI infers a velocity vector field in the space of scRNA-seq data from profiles of individual data, and models the dynamics of cell trajectories with a linear ordinary differential equation to reconstruct the underlying GRN with a sparse regression.
The input to GRISLI is a set of time-stamped scRNA-seq data (xi,ti)i=1,...,C, where C is the number of cells, xi is the vector of GE for the i-th cell and ti is the time associated to the i-th cell; this time can be based on the real experimental time / calculated pseudo-time.

□ xPore: Detection of differential RNA modifications from direct RNA sequencing of human cell lines
>>
https://www.biorxiv.org/content/10.1101/2020.06.18.160010v1.full.pdf
xPore identifies positions of m6A sites at single base resolution, estimates the fraction of modified RNAs in the cell, and quantifies the differential modification rate across conditions.
xPore fits a multi-sample two-Gaussian mixture model, and infer directionality of modification rate differences by utilizing information across all tested positions.

□ CoRE-ATAC: A Deep Learning model for the functional Classification of Regulatory Elements from single cell and bulk ATAC-seq data
>>
https://www.biorxiv.org/content/10.1101/2020.06.22.165183v1.full.pdf
CoRE-ATAC, a deep learning framework with novel data encoders that integrate DNA sequence (reference or personal genotypes) and ATAC-seq read pileups.
CoRE-ATAC integrates DNA sequence data with chromatin accessibility data using a novel ATAC-seq data encoder that is designed to be able to integrate an individual’s genotype with the chromatin accessibility maps by inferring the genotype from ATAC-seq read alignments.

□ PORE-cupine: Direct RNA sequencing reveals structural differences between transcript isoforms
>>
https://www.biorxiv.org/content/10.1101/2020.06.11.147223v1.full.pdf
PORE-cupine, an approach that combines structure probing with SHAPE-like compound NAI-N3, nanopore direct RNA sequencing, and one-class support vector machines to detect secondary structures on near full- length RNAs.
PORE-cupine captures structural information in a transcriptome rapidly and directly. The nature of long-read sequencing through nanopores also allows us to accurately assign and capture structures and their connectivity along individual gene-linked isoforms.

□ DReSS: A difference measurement based on reachability between state spaces of Boolean networks
>>
https://www.biorxiv.org/content/10.1101/2020.06.19.161224v1.full.pdf
Structure perturbation can change the system’s state space from one to another. to evaluate the influence of a specific structure perturbation to the system’s state space, is actually to evaluate the difference between two directed networks.
DReSS, Difference based on Reachability between State Spaces can quantitively describe the changes of reachability of networks’ state spaces.

□ SCIM: Universal Single-Cell Matching with Unpaired Feature Sets
>>
https://www.biorxiv.org/content/10.1101/2020.06.11.146845v1.full.pdf
SCIM assumes that cells share a common (low-dimensional) underlying structure and that the underlying cell distribution is approximately constant across technologies.
SCIM constructs a technology-invariant latent space using an auto-encoder framework with an adversarial objective. Multi-modal datasets are integrated by pairing cells across technologies using a bipartite matching that operates on the low-dimensional latent representations.
□ MMD-MA: Unsupervised manifold alignment for single-cell multi-omics data
>>
https://www.biorxiv.org/content/10.1101/2020.06.13.149195v1.full.pdf
Maximum mean discrepancy manifold alighment (MMD-MA)—that approaches integration of heterogeneous single-cell data sets as an unsupervised embedding problem.
MMD-MA employs an objective function that minimizes the maximum mean discrepancy (MMD) between the data sets in the latent space, while also maintaining the underlying structure of each data set.
Averaging the fraction across all data points in both domains yields the average “fraction of samples closer than the true match” (FOSCTTM), where perfect recovery of the true manifold structure will yield a value of zero.

□ KAML: Improving Genomic Prediction Accuracy of Complex Traits Using Machine Learning Determined Parameters
>>
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-02052-w
KAML, Kinship Adjusted Multiple Loci Best Linear Unbiased Prediction is designed to predict genetic values using genome-wide or chromosome-wide SNPs for either simple traits that controlled by a limited number of major genes or complex traits that influenced by many polygenes with minor effects.
The model parameters are optimized using the information of bootstrap strategy based GWAS results in a parallel accelerated machine learning procedure combing cross-validation, grid search and bisection algorithms.
KAML provides a flexible assumption to accommodate traits of various genetic architectures and incorporates pseudo-QTNs as fixed effect terms and a trait-specific random effect term under the LMM framework.

□ sdcorGCN: Robust gene coexpression networks using signed distance correlation
>>
https://www.biorxiv.org/content/10.1101/2020.06.21.163543v1.full.pdf
sdcorGCN, a framework to generate self-consistent networks using signed distance correlation purely from gene expression data, with no additional information.
sdcorGCN constructs networks by including only edges between genes for which the signed distance correlation of their expression exceeds a threshold based on the internal consistency of the networks COGENT instead of using exogenous biological information known a priori.
□ COGENT: evaluating the consistency of gene co-expression networks
>>
https://www.biorxiv.org/content/10.1101/2020.06.21.163535v1.full.pdf
COGENT - COnsistency of Gene Expression NeTworks, designed to aid the choice of a network construction pipeline without the need for annotation or external data.
COGENT evaluates network construction methods through iterative resampling. COGENT can be used to select between Pearson and Kendall correlation coefficients for measuring co-expression, as well as how to select the score cut-off.

□ Synthetic observations from deep generative models and binary omics data with limited sample size
>>
https://www.biorxiv.org/content/10.1101/2020.06.11.147058v1.full.pdf
There are two potential reasons, why deep Boltzmann machines (DBMs) perform better compared to generative adversarial networks (GANs) at rather small sample sizes and, compared to variational autoencoders (VAEs), generally better learn the magnitude of the signal.
compared to DBMs, VAEs and GANs require to learn more parameters since they rely on feed-forward networks. The second reason is related to the regularization which is applied during parameter optimization.
□ rearrvisr: an R package to detect, classify, and visualize genome rearrangements
>>
https://www.biorxiv.org/content/10.1101/2020.06.25.170522v1.full.pdf
rearrvisr provides functions to identify and visualize inter- and intrachromosomal translocations and inversions between a focal genome and an ancestral genome reconstruction, or two extant genomes.
rearrvisr directly maps rearrangements onto the focal genome, enabling the localization of rearranged genomic regions and facilitating the determination of their extent.
□ ExaStoLog: Exact solving and sensitivity analysis of stochastic continuous time Boolean models
>>
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03548-9
The analysis confirmed the possibility of efficiently applying exact methods in the context of stochastic logical models, as well as the importance of their parametric analysis.
topological sorting of the state transition graph and the dependencies between the nullspaces and the kinetic matrix. Up to an intermediate size stochastic Boolean models can be efficiently solved by an exact matrix method, without using Monte Carlo simulations.

□ PLEIO: A method to map and interpret pleiotropic loci using summary statistics of multiple traits
>>
https://www.biorxiv.org/content/10.1101/2020.06.16.155879v1.full.pdf
PLEIO utilizes an optimization technique using spectral decomposition of the variance.
PLEIO maximizes power by systematically accounting for the genetic correlations and heritabilities of the traits in the association test. Any set of related phenotypes, binary or quantitative traits with differing units, can be combined seamlessly.

□ BAGEA: A framework for integrating directed and undirected annotations to build explanatory models of cis-eQTL data
>>
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007770
Bayesian Annotation Guided eQTL Analysis (BAGEA) integrates directed genomic annotations with eQTL summary statistics from tissues of various origins. BAGEA can be run on summary statistics using external LD information as well as on individual level genotype data directly.
BAGEA can directly model phenomena relevant to genetic architecture, such as the relatively larger impact of SNPs close to the TSS on directed annotations compared to that of distal SNPs. BAGEA can model multiple causal SNPs per region.
□ Metasubtract: An R‐package to Analytically Produce Leave‐one‐out Meta‐analysis GWAS Summary Statistics
>>

https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btaa570/5858976
METAL and MetaSubtract results of genetic markers that were present in every cohort were compared for the corrected effect size, SE, z-score, -log(p-value), allele frequency, and Q statistic using two-way mixed ANOVA intraclass correlation coefficients with absolute agreement.
□ BayesHL: Bayesian Hyper-LASSO Classification for Feature Selection
>>
https://www.nature.com/articles/s41598-020-66466-z
a Bayesian Robit regression method with Hyper-LASSO priors (BayesHL) for feature selection in high dimensional genomic data with grouping structure.
The main features of BayesHL include that it discards more aggressively unrelated features than LASSO, group LASSO, supervised group LASSO, penalized logistic regression, random forest, neural network, XGBoost and knockoff.
□ Nonlinear ridge regression improves robustness of cell-type-specific differential expression studies
>>
https://www.biorxiv.org/content/10.1101/2020.06.18.158758v1.full.pdf
Nonlinear regression, which models scales properly, is recommended more than the linear regression, yet the difference can be modest.
Subsequently, cell-type-specific effects are estimated by linear regression that includes terms representing the interaction between the cell type proportions and the trait. This approach involves two issues, scaling and multicollinearity.
□ Fast Sparse-Group Lasso Method for Multi-response Cox Model with Applications to UK Biobank
>>
https://www.biorxiv.org/content/10.1101/2020.06.21.163675v1.full.pdf
Multi-snpnet-Cox, a Sparse-Group regularized Cox regression method to analyze large-scale, ultrahigh-dimensional, and multi-response survival data efficiently.
A Sparse-Group penalty that encourages the coefficients to have small and overlapping support; A variable screening procedure that minimizes the frequency of disk memory access; An accelerated proximal gradient method that optimizes the regularized partial-likelihood function.
□ ExpResNet: Predicting Gene Expression from DNA Sequence using Residual Neural Network
>>
https://www.biorxiv.org/content/10.1101/2020.06.21.163956v1.full.pdf
ExpResNet, a deep residual network model to predict gene expression directly from DNA sequence.
ExpResNet consists of five residual units, each followed by an adaptive average pooling layer, and two fully connected layers with a batch normalization a layer and a ReLU layer in between the two layers.
□ Gaussian Mixture Model-Based Unsupervised Nucleotide Modification Number Detection Using Nanopore Sequencing Readouts
>>
https://academic.oup.com/bioinformatics/article-abstract/doi/10.1093/bioinformatics/btaa601/5864718
a framework for the unsupervised determination of the number of nucleotide modifications from nanopore sequencing readouts.
It can effectively recapitulate the number of modifications, the corresponding ionic current signal levels, as well as mixing proportions under both DNA and RNA contexts.
by integrating information from multiple detected modification regions, that the modification status of DNA and RNA molecules can be inferred.
□ Nested Stochastic Block Models Applied to the Analysis of Single Cell Data
>>
https://www.biorxiv.org/content/10.1101/2020.06.28.176180v1.full.pdf
As there could be more model fits with similar entropy, schist could explore the space of solutions with a Markov Chain Monte Carlo algorithm, to perform model averaging: that is the difference in model entropy in n continuous iterations remains under a specified threshold.
The computational framework underlying schist calculates the model entropy, that is the amount of information required to describe a block configuration. schist performs an exhaustive exploration of all model entropies resulting from moving all cells into all possible clusters.
□ kTWAS: integrating kernel-machine with transcriptome-wide association studies improves statistical power and reveals novel genes
>>
https://www.biorxiv.org/content/10.1101/2020.06.29.177121v1.full.pdf
kTWAS leverages TWAS-like feature selection followed by a SKAT- like kernel-based score test, to combine advantages from both approaches.
kTWAS will take advantage of TWAS-based feature selection, which is directed by expression data, as well as a kernel-based association test, which is robust to the underlying genetic architecture of the focal phenotype.

□ Compact Integration of Multi-Network Topology for Functional Analysis of
Genes
>>
https://www.cell.com/cell-systems/fulltext/S2405-4712(16)30360-X
Mashup decouples the dimensionality of feature representations from the data parameters, which allows it to cope with inherent noise in high-throughput data by obtaining compact representations that keep only the most explanatory features.
In Mashup, the diffusion in each network is first analyzed to characterize the topological context of each node. Next, the high-dimensional topological patterns in individual networks are canonically represented using low-dimensional vectors.

□ Exploring generative deep learning for omics data by using log-linear models
>>

https://academic.oup.com/bioinformatics/article/doi/10.1093/bioinformatics/btaa623/5869514
an approach that extracts patterns from synthetic samples and corresponding latent representations learned by a deep generative approach such as VAEs or deep Boltzmann machines.
Modeling large contingency tables with log-linear models can be time consuming when more than 10 features are intended to be selected, i.e. the resulting contingency table is at least 11-dimensional.

□ DISC: a highly scalable and accurate inference of gene expression and structure for single-cell transcriptomes using semi-supervised deep learning
>>
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-02083-3
DISC, a novel deep learning network with semi-supervised learning to infer gene structure and expression obscured. DISC integrates an AE and RNN and uses SSL to train model parameters.
DISC employs semi-supervised learning and its loss function is computed on both positive-count genes (real labels) and zero-count genes (pseudo labels). DISC distinguishes the technical zero generated by down-sampling.











 (iPhone 11 Pro. 17/07/2020.)
(iPhone 11 Pro. 17/07/2020.)