
□ The Future of Medicine Is Not In Your Hands (Yet)
>> http://goo.gl/Re1gXQ

□ NewUniverseD:
Astronomers Discover Record Breaking Quasar http://buff.ly/1wuLkqy

□ Scientists Find Fractal Patterns in Variable Stars
>> http://buff.ly/1wA7k3l
The "underlying nonchaotic attractor" is not dark matter and gravity.

□ Cubist Saturn
>> http://astronomynow.com/2015/02/24/cubist-saturn/
The view was obtained at a distance of approximately 2 million kilometres (1.2 million miles) from Saturn. Image scale is 11 kilometres (7 miles) per pixel.

□ The 16th annual Advances in Genome Biology and Technology (AGBT)
>> http://www.agbt.org
>> #AGBT15
□ 10X Genomics at AGBT:
>> http://www.bio-itworld.com/2015/2/25/10x-genomics-agbt.html
10X genomicsの技術摘要。the GemCode Platformと命名されたらしい。DNAサンプルを14塩基分子のバーコードを割り当て断片化。Matrix Viewによる可視化。
at AGBT, the 10X technology finally has a name: the GemCode Platform, including an instrument, chemistry kit, and informatics software. The DNA is fragmented into short-read libraries suitable for Illumina and receiving a 14-base molecular barcode unique to its gem of origin. The GemCode Software works on “Linked Read Data ” where every short read can be binned back to its DNA molecule of origin.
>> https://vimeo.com/120429438
□ RM, using BWA-MEM + Lumpy / Localize HGAP + CA + Mummer / Whole genome assembly (done by DNAnexus in less than one day)
□ WRM: "World's fastest genome assembly" in 22hrs using FALCON+Daligner on @dnanexus. #PacBio #SMRTseq
今年のAGBTに関するコメントで「10x Genomicsの発表の前に『オ・フォルトゥーナ』を演奏すべきだ」に笑うのと同時に、新プラットフォームへの現地の期待感がよく伝わる。
□ Linked Read Algorithms for Haplotype Phasing and Structural Variant Detection
□ CIViC database for clinical interpretation of variants in cancer, open source & community-driven:
>> https://civic.genome.wustl.edu/
□ obigriffith:
#CIViCdb provides a fully open source and open access solution to knowledge curation bottleneck for precision medicine. #AGBT15
□ CIgenomics:
#AGBT15 software demos were good; @GenomOncology @GenoLogics@obigriffith with CIViC; and finally
@scilifelab almost ready for 2D RNA-seq
□ Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line:
>> http://genome.cshlp.org/content/24/12/2059.full
□ PacBio:
Seeing the Genome in a New Light (Sunshine?) with SMRT Sequencing
>> http://blog.pacificbiosciences.com/2015/02/agbt-2015-seeing-genome-in-new-light.html?m=1 …

gene changes in space
□ LM: Genomic portrait foa space neuron, some of the abundant transcripts in neurons are non-coding RNAs, but down-regulated in space #AGBT15
Leonid Moroz: "Neurons evolved more than once, 9-12 ways to make a brain. Access 3.5 billion years of experiments already done in evolution"
宇宙における無重力環境でのゲノムへの影響をトラッキングするSpace-Seqという概念。シーケンシング原理が、塩基よりMolecular-Basisに偏って行けば、例えば地球外の未知生命が発見された場合の遺伝子的レベルでの相互毒性を計る指標にもなるかもしれない。
□ Evan Macosko to present "Drop-Seq", transformative new single-cell seq technology, at 3 pm at #AGBT15. Not yet tested at zero-gravity tho.
□ Drop-Seq: Single Cell RNA-Seq on a Massive Scale using DNA Barcode Beads and Droplet Microfluidics:
>> http://weitzlab.seas.harvard.edu/research/current/oni-basu …
□ G&T-seq: Separation and parallel sequencing of the genomes and transcriptomes of single cells
G&T-seq provides whole genome amplified genomic DNA and full-length transcript sequence and with automation, 96 samples can be processed in parallel.
□ iGenomics: Alignment and variant calling on the iPhone.
>> http://schatzlab.cshl.edu/iGenomics/tutorial/
iPhoneでIllumina, Ion Torrent, PacBio, MinIONのSeqデータを扱える

□ GenoPharmix is offering free visualizations for your data based on d3.
>> http://genopharmix.com/genopharm-v0.2/solutions.html
Deep Learningアルゴリズム "Tuatara GS1"による関係抽出メソッド、Cognitive Biomimicryを応用したソリューションや、テキストマイニング、可視化による知見提供。実験的サービスではあるけど、アウトソーシングビジネスとして成立するか注視したい。
Deep LearningアルゴリズムをGraph Serverと紐付けて解析するサービス、ゲノム分野でも主流となる可能性がある。ピンカーの理論に依る処の、人の認知不可能なレベルの複雑なデータからの関連性抽出は仮説生成に有用。ただし収益性のあるハードとして提供出来るかどうかは課題
In silico emulation of biomimetic object-association strategies has proven to be very effective in relationship discovery and deep learning, leading to new hypothesis and new discoveries based on pushing the boundaries of data science. the algorithm set accomplishes is similar to pLSI or probabilistic latent semantic indexing aka probabilistic latent semantic analysis PLSA.
The methods & algorithm sets were primarily forged for Life Sciences work in the area of Genomic pathway prediction/analysis at Berkeley Labs.
probalistic approaches in high-dimensional vector space designed for the purpose of mimicking portions of human cognition principles via CTM.
Biomimetic high-dimensional vector space algorithm sets that generate relationships between objects are based on CTM and auto-association.

□ Investigation of gene-gene interactions in dose-response studies w/ Bayesian nonparametrics:
>> http://www.biodatamining.org/content/pdf/s13040-015-0039-3.pdf …
MANOVA and the novel Bayesian framework present a trade-off between computational complexity and model flexibility. Bayesian posterior probabilities are computed to assess how likely each SNP is to be involved in determing drug-response.
the Bayesian neural network can be employed on a smaller subset of SNPs to explore a richer model space.
As with any MCMC method, sub-optimal parameter values will result in a chain failing to converge, even in Bayesian neural network framework.

□ Differential co-expression network centrality and machine learning feature selection for identifying susceptibility hubs in networks with scale-free structure
>> http://www.biodatamining.org/content/pdf/s13040-015-0040-x.pdf
a novel simulation strategy generates microarray case–control data w/ embedded differential co-expression networks. and underlying correlation structure based on scale-free or Erdos-Renyi (ER) random networks.

□ Quantitative and logic modelling of molecular and gene networks:
>> http://www.nature.com/nrg/journal/v16/n3/full/nrg3885.html
a hybrid approaches will become essential for further progress in synthetic biology and in the development of virtual organisms.
Network inference methods: The four main approaches to infer networks from data include correlation (part a), information theoretic (part b), Bayesian inference (part c) and differential equations (part d).
□ Prometheus Project: integrated data platform that enables the world to fulfill the promise of genomics for medicine
>> https://www.broadinstitute.org/dsde/
□ infoecho:
I am always curious about the relationship between "Better machine learning" and "Better learning about machines" :)
□ Data intensive biology and data provenance graphs - Gianluigi Zanetti:
>> http://youtu.be/3Ftd3NBzaZ8
"Actionable" data provenance graph.
・Full tracing of the graph of operations performed.
・Large scale (10^3 genomes) comparisons usage.

□ tranSMART_org:
In town for #TRICON? Come see us at #booth115 for the latest on the tranSMART platform for translational research.
>> http://transmartfoundation.org
□ tranSMART: A collaborative approach to develop a multi-omics data analytics platform for translational research
>> http://www.sciencedirect.com/science/article/pii/S2212066114000350
To fully utilize the strengths of tranSMART, several functionalities are required that can be complemented by the strong algorithmic capabilities included in Genedata Analyst.
□ Life Science and IT Organizations Invest in the tranSMART Foundation to Advance Translational Medicine Research:
>> http://www.marketwatch.com/story/life-science-and-it-organizations-invest-in-the-transmart-foundation-to-advance-translational-medicine-research-2015-01-14 …
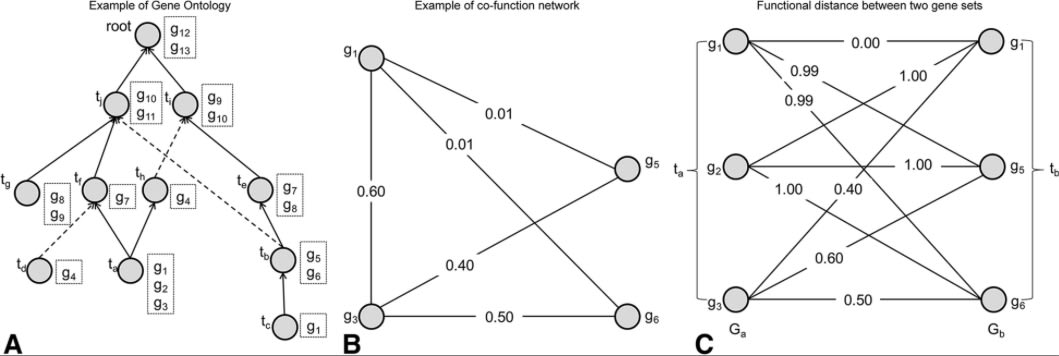
□ Measuring semantic similarities by combining gene ontology annotations and gene co-function networks:
>> http://www.biomedcentral.com/content/pdf/s12859-015-0474-7.pdf …
NETSIM calculates the functional distance between a pair of gene sets that are annotated to a pair of GO terms. Second, it calculates GO term similarity based on the annotations to the common parent term, but propagates only the annotations to the terms that lie on the paths from the two GO terms to the common parent term.
Ga ∩ Gb ≠ ∅, and U(ta, tb, p) = Gp. Therefore, D(ta, tb) = 1 and S(ta, tb, p) = 2IC(p)/(IC(ta) + IC(tb)) × (1 - |Gp|/|G|)

(different enrichment measures on a small region of GO DAG. rectangles contain subset of genes annotated by each node)
□ A Bayesian extension of the hypergeometric test for functional enrichment analysis:
>> http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3954234/
a Bayesian approach based on the non-central hypergeometric model to addresses the limitations of the traditional hypergeometric P-value. The Gene Ontology dependence structure is incorporated through a prior on non-centrality parameters.
Gene Ontologyの超幾何分布モデルに対するベイズ・アプローチ。尤度関数には重複情報を含まない。
□ Patent: Long fragment de novo assembly using short reads
>> http://www.freepatentsonline.com/y2015/0057947.html
Complete GenomicsのRadoje氏の取得した特許。あれ?これって。。
A kmer index can include labels indicating an origin of each of the nucleic acid molecules that include each kmer, memory addresses of the reads that correspond to each kmer in the index, and a position in each of the mate pairs that includes the mer.
□ Correcting Illumina sequencing errors: extended background:
>> http://lh3.github.io/2015/02/13/comments-on-illumina-error-correction/ …
>> http://arxiv.org/pdf/1502.03744v1.pdf
Input: K-mer size k, set H of trusted k-mers, and one string S
Output: Set of corrected positions and bases changed to
Function CORRECTERRORS(k,H, S) begin Q←HEAPINIT()
HEAPPUSH(Q,(k - 2, S[0, k - 2], ∅, 0))
while Q is not empty do
(i, W, C, p)←HEAPPOPBEST(Q)
i←i+1
if i = |S| then return C
N ← {(i,A),(i,C),(i,G),(i,T)}
□ Improved data analysis for the MinION nanopore sequencer:
>> http://www.nature.com/articles/nmeth.3290.epdf …
Over 99% of high-quality 2D MinION reads mapped to the reference at a mean identity of 85%.
□ Minimum Information for Reporting Next Generation Sequence Genotyping (MIRING):
>> http://biorxiv.org/content/early/2015/02/16/015230 …
The data recorded in a MIRING message are essential for the systematic traceability of a NGS genotyping result. MIRING incl 5 categories of structured information: message annotation/reference context/full genotype/consensus sequence/novel polymorphism

□ Deep Neural Nets as a Method for Quantitative Structure–Activity Relationships:
>> http://pubs.acs.org/doi/abs/10.1021/ci500747n …
□ Multi-task Neural Networks for QSAR Predictions: http://arxiv.org/abs/1406.1231
Merckのコンペティションで1st placeを受賞した技術を応用したものらしい。
neural nets with multi-tasking can lead to significantly improved results over baselines generated with random forests. Neural networks are powerful non-linear models for classification, regression, or dimensionality reduction.
The most direct application of neural networks to QSAR modeling is to train a neural net on data from a single assay using vectors of molecular descriptors as input and recorded activities as training labels.

□ Architecture of the DNN used to predict AS patterns: Deep learning of the tissue-regulated splicing code (MKK Leung)
It Contains 3 hidden layers, with hidden variables that jointly represent genomic features and cellular context.
□ Deep Learning: Learning deep representations for single cell genomics: http://www.ndm.ox.ac.uk/principal-investigators/project/deep-learning-learning-deep-representations-for-single-cell-genomics … Nuffield Department of Medicine
□ GraphSAW: web-based system for graphical analysis of drug interactions and side effects using pharmaceutical and molecular data
>> http://www.biomedcentral.com/1472-
using GraphSAW which analyzes multi-medications and evaluates with regards to pharmaceutical and molecular adverse drug reactions.
□ DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis:
>> http://bioinformatics.oxfordjournals.org/content/31/4/608.abstract …
Enrichment analyses incl hypergeometric model and gene set enrichment analysis are implemented to support discovering disease associations.
□ DIME: A Novel Framework for De Novo Metagenomic Sequence Assembly:
>> http://online.liebertpub.com/doi/abs/10.1089/cmb.2014.0251
メタゲノムアセンブリにMapReduceを実装をすることで、理論速度に近いシーケンスを可能に。
two MapReduce implementations of DIME, DIME-cap3 and DIME-genovo, tested comparison of Cap3, Genovo, MetaVelvet, SOAPdenovo, and SPAdes.
a multilevel k-way graph partitioning algorithm, KMETIS, performs best when the number of parts is more than 8 as stated in the literature.
□ RAPTR-SV: a hybrid method for the detection of structural variants:
>> http://bioinformatics.oxfordjournals.org/content/early/2015/02/16/bioinformatics.btv086.short …
RAPTR-SV had superior sensitivity and precision, as it recovered 66.4% of simulated tandem duplications with a precision of 99.2%.
□ The long road from Data to Wisdom, and from DNA to Pathogen:
>> http://microbe.net/2015/02/17/the-long-road-from-data-to-wisdom-and-from-dna-to-pathogen/
The two most important aspects relevant to public health are that these DNA fragments, “if truly present,” are present at extremely low levels, and that there are no reported cases of either of these pathogens.

(Overview of the flow of the StringTie algorithm, compared to Cufflinks and Traph. )
□ StringTie enables improved reconstruction of a transcriptome from RNA-seq reads:
>> http://www.nature.com/nbt/journal/vaop/ncurrent/full/nbt.3122.html …
StringTie, a computational method that applies a network flow algorithm originally developed in optimization theory.
StringTie produces more complete and accurate reconstructions of genes and better estimates of expression levels, compared with other leading transcript assembly programs including Cufflinks, IsoLasso, Scripture and Traph.

□ Deterministic Chaos and the Evolution of Meaning:
>> http://bjps.oxfordjournals.org/content/63/3/547.full
決定論的カオスと意味の進化: 非収束適応ダイナミクスとゼロサム・シグナリング・ゲーム
a new explanation for the evolution or spontaneous emergence of meaning: non-convergent adaptive dynamics. a zero-sum strategic interaction―information transmission is sustained indefinitely by the replicator dynamic. The key to the persistence of this out-of-equilibrium information transfer is deterministic chaos.
□ SNN-Cliq: a novel algorithm that clusters single cell transcriptomes:
>> http://bioinformatics.oxfordjournals.org/content/early/2015/02/10/bioinformatics.btv088.abstract …
SNN-Cliq utilizes the concept of shared nearest neighbor that shows advantages in handling high dimensional data. to define similarities between data points (cells) and achieve clustering by a graph theory-based algorithm.





















фитнес браслет mi band 3
фитнес браслет mi band 3