□ Sensitive measurement of single-nucleotide polymorphism-induced changes of RNA conformation: application to disease studies
>>
http://nar.oxfordjournals.org/content/early/2012/11/02/nar.gks1009.long
In probability theory and information theory, the ‘relative entropy’ or the ‘Kullback?Leibler divergence’ is an accepted measure of the difference between two probability distributions.
We designed a novel efficient dynamic programming algorithm remuRNA to compute the relative entropy between the wild-type and mutant ensembles. We present the mathematical relationships on which our dynamic program is built in the following subsection.
野生型および変異型RNA構造アンサンブルの相対的エントロピー計算のための動的プログラミング・アルゴリズム
□
fukunagaTsu:
RNAの塩基変化により、構造の分布がどう変化したかのKL-divergenceを求めるアルゴリズムの開発。#Everydayペーパー
Clarity LIMS, Intuitive Lab Management Software for Regulated and Research Labs. from GenoLogics on Vimeo.
□ Clarity LIMS: NGS特化型のラボマネージメントソフトウェア:
Intuitive Lab Management Software for Regulated and Research Labs.
>>
http://www.genologics.com/claritylims
Clarity LIMS is built for NGS laboratories that are CLIA certified or aiming to become certified. Clarity LIMS increases user adoption through exceptional usability and will assist clinical lab directors to ensure adherence to their lab’s SOPs through workflow enforcement.
□
origenebio:
Do you know what Gene Therapy is?
pic.twitter.com/hfHOJWnK

□ 『Rによる計算機統計学 (Statistical Computing with R by Maria L. Rizzo)』
>>
http://ssl.ohmsha.co.jp/cgi-bin/menu.cgi?ISBN=978-4-274-06830-0
購入したよo(*゜▽゜*)o モンテカルロ法を中心とした解析法のかなり実践的な内容。ざっと見、翻訳も評判より悪くない p
ブートストラップ、ジャックナイフでのモンテカルロ法、パーミューテーションテスト(並べ替え検定)、マルコフ連鎖モンテカルロ(MCMC)法などの高度な統計手法を用いた解析についてきちんと解説し、その使用法を示したもの。解析についてはR により、統計学的コンピューティングの事例によるアプローチで計算統計の古典的な主要問題を解説した。全事例にR のコードを付け、R 言語のプログラミングの概念の説明を補足するようにした。
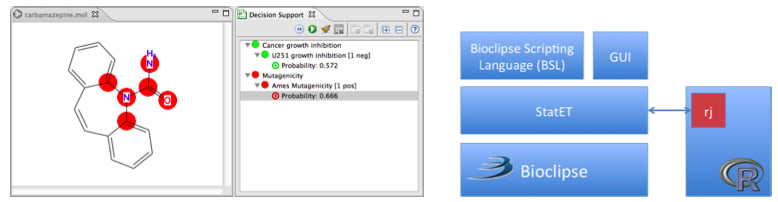
□
egonwillighagen:
ah, our Bioclipse-R paper is online!
>>
http://bioinformatics.oxfordjournals.org/content/early/2012/11/23/bioinformatics.bts681.short
Bioclipse-R: Integrating management and visualization of life science data with statistical analysis
>>
http://m.bioinformatics.oxfordjournals.org/content/early/2012/11/23/bioinformatics.bts681.full.pdf
Bioclipse, a graphical workbench for the life sciences, provides functionality for managing and visualizing life science data. We introduce Bioclipse-R, which integrates Bioclipse and the statistical programming language R. The synergy between Bioclipse and R is demonstrated by the construction of a decision support system for anticancer drug screening and mutagenicity prediction, which shows how Bioclipse-R can be used to perform complex tasks from within a single software system.
□ GWAS Diagram Browser: automatic data-driven generation of the GWAS diagram / iconic visualisation of the catalogue data
http://www.ebi.ac.uk/fgpt/gwas/
□ The birth of the Epitranscriptome: deciphering the function of RNA modifications
>>
http://genomebiology.com/2012/13/10/175/abstract
Recent studies have found methyl-6-adenosine in thousands of mammalian genes, and this modification is most pronounced near the beginning of the 3' UTR. We present a perspective on current work and new single-molecule sequencing methods for detecting RNA base modifications.
□
BiotechGuide:
2 Biotech Companies To Consider In The Wake Of Positive Developments
http://sns.mx/YmiLy7
□
nygenome:
Congratulations to @
Stanford @
PhxChildrens & @
UConn with @jacksonlab on their new genomic research centers!
http://bit.ly/UfkhMB
□
stem_cells:
Video protocol: How to isolate PBMCs from whole #blood in just 15 minutes
http://ow.ly/fVbvR #immunology
□ RNAi and Antisense Targeting the Same Gene: Not a Zero-Sum Game: The TTR Amyloidosis Race: Alnylam - ISIS battleground
>>
http://rnaitherapeutics.blogspot.de/2012/11/demystifying-ddrnai-trigger-design.html
Transthyretin-mediated amyloidosis has become the single-most important factor for the 3-5 year valuation of certainly Alnylam, and possibly ISIS Pharmaceuticals as well. After TTR, Z-alpha-1-antitrypsin (Z-AAT) is gearing up as the next Alnylam(RNAi)-ISIS(antisense) battleground. Expect to hear competitive language from the two camps why their approach will prove superior over the other. It is, however, worth keeping in mind that having two candidates race towards approval can also have significant pie-enlarging benefits.
Alnylam社とISIS医薬品による、トランスサイレチン(TTR)アミロイドーシス治療を主戦場とした競争。最初に商業的成功を収めるのは、RNAi TherapeuticsかRNaseH antisenseか

□
idtdna: Integrated DNA Tech
Oligo Modification?Post-Synthesis Conjugation Explained
>>
http://ow.ly/fCT9Q
Modifications can be incorporated into synthetic oligonucleotides in a variety of ways. Standard DNA bases are synthetically coupled via phosphoroamidite chemistry. The reaction proceeds in the 3’ to 5’ direction where the 5’ hydroxyl group of each base attaches to the 3’ phosphate group of the next base. Many modifications, such as 6-FAM, standard biotin, and internal Cy3, can be attached via phosphoroamidite chemistry directly on the synthesis column
□
Stephen Turner ?:
metAMOS: assembly and analysis toolkit for #metagenomics
http://www.cbcb.umd.edu/software/metAMOS … code:
https://github.com/treangen/metAMOS/wiki … VM:
https://gembox.umiacs.umd.edu/data/public/e6ba231f76296c8f5e37aba5bbb234f2.php …
metAMOS is an integrated assembly and analysis pipeline for metagenomic data. It is built around the Bambus2 metagenomic scaffolder and includes many current tools for assembly, gene finding, and taxonomic classification.
□
hinaichigo:
MetaVelvetとかSparseAssemblerとかGlimmer-MG入れてるとか良いカスタマイズね。
□
moorejh:
Nice Fig 1 "The eukaryotic #genome as an #RNA machine" Mattick
http://www.ncbi.nlm.nih.gov/pubmed/18369136 #genomics
#HMRIpmw #complexity #epigenomics @myen
□
_frankie_ann:
"RNA as substrate for epigenome-env interactions" Context dependent interaction of RNA w/ gene expression. #HMRIpmw #johnmattick
□
_frankie_ann:
Beware hypothesis driven research and the inevitable resultant false positives... #HMRIpmw #danielmacarthur
□
jcfournet:
GEN | Insight & Intelligence™: Open Innovation in the Pharma Industry
http://lnkd.in/YNqSDT

□
Matt_Landau:
For data mining and semantically-enriched searching, @BioMedCentral's Cases Database launches today -
>>
http://www.casesdatabase.com/
Cases Database, a continuously-updated, freely-accessible database of over 10 000 medical case reports from multiple publishers, including Springer, BMJ and PubMed Central.
□
BMC_series:
Embrace “information overload” with Cases Database - @BioMedCentral blog
http://blogs.biomedcentral.com/bmcblog/2012/12/10/embrace-information-overload-with-cases-database/ … #openaccess #oa
□
chronicle:
NIH considers allowing anonymity for grant applications:
http://bit.ly/TPGwwd
□
yokofakun:
My NGS workflow (16 exomes) visualized in #gephi.
pic.twitter.com/8ZQ6G37I
□ "Don't Reinvent the Wheel" Ten Simple Rules for the Open Development of Scientific Software:
>>
http://t.co/WFKqmCDC
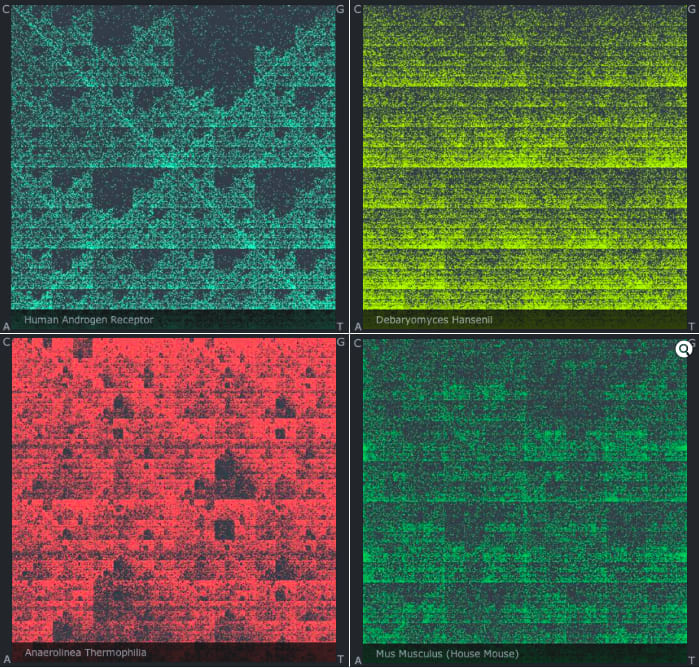
□ Using Chaos to Visualize DNA Sequences:
>>
http://www.itsokaytobesmart.com/post/36683803258/chaos-dna-sequences
Comparing two enormous sequences is very difficult too. Say you want to see the patterns or differences that exist between the human and chimpanzee genomes. Some of the world’s most powerful supercomputers are required in order to meaningfully align such long strings of DNA bases, letter by letter, piece by piece.
□ Games With DNA - new method of comparing graphical representations of DNA sequences:
>>
http://dl.dropbox.com/u/18166613/cgr/cgr_description.html …
Chaos Game Representation (CGR)
a powerful method for graphical representation and analysis of DNA. It creates a scatter plot derived from a DNA sequence. Each of the points on the plot corresponds to one base of the sequence, A, C, T, or G. The first nucleotide of the sequence is plotted halfway between the center of the unit square and the corresponding corner of the nucleotide. The next point is plotted halfway between the next corresponding corner and the previous point. This cycle continues until all nucleotides of the sequence have been read, and the sequence has been plotted.
ゲノム配列の相違をフラクタル・パターンで可視化・比較する"CGR"
□
isatools:
Peter Li: GigaDB and Galaxy - revolutionizing data dissemination, organization … by @gigascience, uses @isatools
http://www.slideshare.net/GigaScience/peter-li-gigadb-and-galaxy-revolutionizing-data-dissemination-organization-and-analysis-15382102 …
□
appliedgenomics:
Stellar defense of NIH funding mechanism from Jeremy Berg, but I doubt I'm alone in thinking the system needs to change
http://scopeblog.stanford.edu/2012/12/05/nih-funding-mechanism-totally-broken-says-stanford-researcher/ …
□ Bioinformatics in the cloud: Biology Direct review covers cloud based resources and opportunities
>>
http://www.biology-direct.com/content/7/1/43/abstract … #bioinformatics #
ICG7
Albeit relatively new, cloud computing promises to address big data storage and analysis issues in the bioinformatics field. Here we review extant cloud-based services in bioinformatics, classify them into Data as a Service (DaaS), Software as a Service (SaaS), Platform as a Service (PaaS), and Infrastructure as a Service (IaaS), and present our perspectives on the adoption of cloud computing in bioinformatics.
□
NCBI:
SRA contains >10^15 bases (a PetaBase!) of data w/ 390TB of open-access data & 610TB of controlled-access clinical data.http://1.usa.gov/RztMKU
□
appliedgenomics:
Biobase to Market ANNOVAR Annotation Tool - kudos to Kai Wang, who developed the tool while at our Lab!
http://www.genomeweb.com/informatics/biobase-market-annovar-annotation-tool#.UL92WVAb2mU.twitter …
□ Developments in next generation sequencing ? a visualisation:
>>
http://flxlexblog.wordpress.com/2012/12/03/developments-in-next-generation-sequencing-a-visualisation/ …
□ DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs:
>>
http://m.nar.oxfordjournals.org/content/early/2012/11/28/nar.gks1246.full
>>
www.microrna.gr/LncBase
DIANA-LncBase hosts transcriptome-wide experimentally verified and computationally predicted miRNA recognition elements (MREs) on human and mouse lncRNAs. The analysis performed includes an integration of most of the available lncRNA resources, relevant high-throughput HITS-CLIP and PAR-CLIP experimental data as well as state-of-the-art in silico target predictions.
The experimentally supported entries available in DIANA-LncBase correspond to >5000 interactions, while the computationally predicted interactions exceed 10 million. DIANA-LncBase hosts detailed information for each miRNA?lncRNA pair, such as external links, graphic plots of transcripts’ genomic location, representation of the binding sites, lncRNA tissue expression as well as MREs conservation and prediction scores.
□
multiscalebio:
Mount Sinai's Genomic Training for Doctors Gets Personal
http://nygenome.org/blog/mount-sinais-genomic-training-doctors-gets-personal … via @
nygenome
Institute of Genomics and Multiscale Biology.
□
dnanexus:
Central theme from #ASHG2012? Responsibility. Check our blog for highlights.
http://ow.ly/ftGc0
□
KaitlanChu:
@IBM @ClevelandClinic collaborate to design Medical Search Engine capable of human-like communication
http://ow.ly/fm7DJ #hci #himss #ux
□
emblebi:
Our RNA Genomics group is looking for a scientific software developer or progammer to develop data analysis interfaces:
http://bit.ly/WIQLED
□
carlzimmer:
@leonidkruglyak @SamWangPhD What I wonder is, what advances in genomics & AI are newsworthy? Promethean revolutions or modest increments?
□ NextGen, Microsoft team up to launch medication app:
>>
http://www.mhimss.org/news/nextgen-microsoft-team-launch-medication-app …
□
scilogscom:
Life performs computation much more than you’ve ever thought. By @MaliciaRogue
http://goo.gl/4tO4D #scilogs
□
MaliciaRogue:
Creating the true citizen scientist | @scoopit
http://sco.lt/6NKsm9 A great read from @SciLogs.com 's 'Beyond the Lab'
□
MaliciaRogue:
Peta, Exa, Yotta And Beyond: Big Data Reaches Cosmic Proportions [Infographic] by @TheTechScribe
http://readwrite.com/2012/11/23/peta-exa-yotta-and-beyond-big-data-reaches-cosmic-proportions-infographic … via @RWW
□
aulifescientist:
Victoria Prize winner, Prof. Terry Speed, says bioinformatics needs a clear career path to guarantee healthy future:
http://ow.ly/fyOls
□
BioWorld:
Pharmas seek innovation, partnering strategies to overcome fragmented marketplaces, shrinking pipelines:
http://bit.ly/TqkHlO
□
SpringerOpen:
Interesting reading: "Open access to knowledge will boost Africa’s development"
http://ow.ly/fBtad #openaccess #oa
□
FAOcomdev:
Call for proposals for Research Grants on Innovative Application of ICTs in Addressing Water-related Impacts of #CC
http://ow.ly/fr8fo
□
VirologyJ:
Published: Development of a SYBR green real-time PCR method for rapid detection of sheep pox virus
http://ow.ly/2tqvAy
□
AIP_Publishing:
Complex impedance, responsivity and noise of transition-edge sensors: Analytical solutions for two- and...
http://ow.ly/fY7ME #AIP_ADV
□
WIREsRNA: miRCURY™ RNA Isolation Kits - Biofluids
Exciting new product notice from Exiqon
http://bit.ly/YmYhWh
□
DailyNewsGW:
NanoString Raises $15.3M in Private Financing Round
http://bit.ly/THoMPS
□
yuifu:
Cell - Deciphering and Prediction of Transcriptome Dynamics under Fluctuating Field Conditions
http://www.sciencedirect.com/science/article/pii/S0092867412013529 … //気象データからのイネの遺伝子発現予測の元論文
□
dritoshi:
3W2III-7 12月13日 17:43 第2会場 データベースと機械学習を利用した新しい 1細胞RNA-seq 法とそのデータ解析の開発 http://goo.gl/KNE4n #mbsj2012 よろしくです。
□
nikkei_btj:
Vanderbilt-Ingramがんセンター、次世代シーケンサーで難治乳がんの分子標的を洗い出し: 米国Vanderbilt-Ingram Cancer Centerのresearch faculty、Justin M.... http://nkbp.jp/VHJ1CR
□
elsevierjp:
間もなく開始16時~。午前中は質問も沢山いただきましたRT @elsevierjp: 図書館員の皆様向)世界の著者識別子ORCIDとScopusの連携ツール※ このツールはScopusのご契約がない機関の方でも利用可能です。http://ow.ly/1PSk5T
□ Dark Matter Will Soon Be Detected, Particle Physicists Predict:
>>
http://www.huffingtonpost.com/mobileweb/2012/11/27/dark-matter-particle-physicists_n_2197072.html
□
ArabNetME:
Social Media Use in Arabia [Infographic]
http://ow.ly/fYfGi
□
IDreamofCongo:
Excellent article by @ReutersAfrica Congolese army in #Goma
http://www.reuters.com/article/2012/12/10/us-congo-democratic-army-idUSBRE8B90AQ20121210 … #DRC
□
chihirow:
そういえば、今日のBiosupercomputing Symposiumのプログラム内容。スライドとかあるので、バイオシミュレーションについて興味が有る方は。【PDF】 / “4th-BSCS-proceedings.pdf” http://htn.to/zUNRfY
□ □ ISLiM国際シンポジウム: 4th Biosupercomputing Symposium
― International Symposium for Next-Generation Integrated Simulation of Living Matter (ISLiM)
>>
http://www.csrp.riken.jp/4thbscs/top.html
次世代生命体統合シミュレーションソフトウエア最終年度講演
スーパーコンピュータ「京」をターゲットとして、ライフサイエンス分野のグランドチャレンジングな一貫ソフトとして、分子・細胞・臓器・全身・脳に亘るシミュレーションソフトと、データ・解析融合手法 及び 生命体基盤ソフトウエアを2006年10月から5年あまり6グループ百数十名(?)で開発してきた成果が発表されます。(今年が最終年度)合わせて、海外からこの分野の先端研究グループの指導的研究者9名の講演があります。
□
simplystats:
The statisticians at Fox News use classic and novel graphical techniques to lead with data
http://simplystatistics.org/?p=623
□
yag_ays:
Machine Learning Advent Calendar 2012の2日目を書きました!/PRML 11章 二変量正規分布からのギブスサンプリング #PRML #R - Qiita by @yag_ays on @Qiita_jp
http://qiita.com/items/5bde6addf228b1fe24e6 …
□
gaou_ak:
Xeon Phi、おそらく来年と再来年のISMBまわりで徐々にBioinformaticsでのアプリケーションが見えてくるんだろうし、MICアーキテクチャ自体には結構期待しているが、バイオインフォマティクスにおける深刻なノイマンボトルネックはMICではより深刻なはず。
□
mkasahara:
プログラムなんて先に送っておけばいいので気にしないんですが、Xeon Phi のリングバスはたいていのバイオインフォのアプリでボトルネックになるでしょうね。メモリ帯域使うアプリ多いし。
□
marimiya_clc:
ロブスターアルゴリズムって結構いけてるね。エントロピー使ってリードの中にSTR含むものを選択して、繰り返し領域はあえてマップせず、その両脇の配列を特定して、その距離から繰り返し数を特定してるのか。ただSTRより長い繰り返しがあるとき、エントロピーがどうなるのか気になった。
□
emblebi:
Welcome teachers! This year's #learninglab, "Biology 2.0 - making sense of biological data" has just kicked off.
http://bit.ly/UM6dM2
□
yag_ays:
マーケットプレイスで高騰してる... / 「Rによる医療統計学」はRの使い方と統計解析の基礎が学べる良書(ただし入手困難) - Wolfeyes Bioinformatics beta http://yagays.github.com/blog/2012/11/29/review-introductory-statistics-with-r/ …
□
KamiMasahiro:
日本の医師不足のシミュレーション結果がプロス・ワン誌に掲載されました。情報工学専門家の井元清哉先生らとの共同研究です。
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0050410 …
□
elsevierjp:
RT @myuuko 嫌悪の原因となるものが、知り合いと関連しているときより、知らない人と関連しているときに、嫌悪の感情は強く(実験1)、心拍は低下し(実験2)、回避行動が素早くなる(実験3)。感染症回避の観点から考察。 http://ow.ly/1PSlvY
□
tsuyomiyakawa:
胎児の細胞が脳血液関門を越えて母親の脳に入って増殖するマイクロキメラ現象はマウスの研究でわかっていた。ヒトでも男性しかない遺伝子が成人女性の脳にあったとのこと。 母親は生物学的に子供との(少しではあるが)キメラになっているということか。 http://news.sciencemag.org/sciencenow/2012/09/bearing-sons-can-alter-your-mind.html#.ULp9Z2nbYPQ.twitter …
□
iNut:
BLASTとは生命科学において多くの研究者から信仰を集めている神の名前である。研究者が供物として自身の研究データを差し出すと「奇跡」よって論文に記載すべき謎の文字列を与える。なお、奇跡と呼ばれるだけあって何をしているのか分からないので、信者は何も考えずただデータを差し出せばよい。
□
dojin_tw:
スウェーデン統計局の地域データ可視化サイトのレベルが半端ない
http://www.scb.se/Kartor/Statistikatlas_42_KN_201206/index.html#story=0 …
□
OxfordJNLsJapan:
【OUP BLOG】人為的操作された人権: Journal of Human Rights Practice によるKony 2012に対する4つのレビュー
http://oxford.ly/TINmms
□
ytb_at_twt:
結局みずもと先生は、カントールの対角線論法の議論を「救う」ため、トップオラクルを持つ二階算術は対角線論法が適用できないウィトゲンシュタイン的理論のモデルとなっていると主張したいのだろうか。構成主義者の「計算可能実数は可算個しかない」と言う主張を退けておいて、大して変わらない主張。
で、ウィトゲンシュタイン的な数学の哲学の有益さに関する個人的の疑問は、それが静的で浅い相対主義で「いくつもの異なるアプローチがあり得る」とただ言うだけのように見えてしまうことです。だからなんだ。異なるアプローチが並列する現状、そしてそれら間の複雑なインタラクションを捉えていない。
□
tri_iro:
(5) 水本さんのトップオラクルモデルじゃ上手く行ってなくね? ちょっと色々調べてみよう。 ⇒ (6) おお、無限時間チューリング計算可能なものだけからなるトポスを考えると、実数から自然数への単射がある!!すげーー!! ⇒ (7) 完。
□
shun0102:
なるほど、15台クラスタ構成で1億レコードではimpalaはhiveの8倍くらいの性能だったけど、10億レコードではスワップ多発で2倍程度になったとのこと。これでImpalaの現状の性能はだいぶわかった感がある http://www.atmarkit.co.jp/ait/articles/1212/07/news011.html …
□
itokouta:
@syuichiao @DrMagicianEARL 「星座」といえばISIS-2試験。Lancetはサブグループ解析を要求。解析に懐疑的な筆者は、双子座と天秤座の患者さんにアスピリンの二次予防効果が認められなかったことを公表して反撃。http://www.ncbi.nlm.nih.gov/pubmed/2899772
□
DrMagicianEARL:
あなた具体的に虚偽を指摘されたら指摘してきた相手をブロックしてますよね @mariscontact 反原発連合のこの方、よく私にいちゃもんつけてくるよ。具体性もなく、虚偽と言い →野間易通@kdxn エートスについて竹野内真理が流している情報のほとんどは虚偽。
反原発派がするべきことは第一に、声が大きいけど資質に問題のある(←ソフトな言い回し)オピニオンリーダーを引きずり降ろすことに尽きる。ツイッターなんかでちやほやして音頭とってる場合じゃない。自分たちの首を締めてるだけ。
□
PKAnzug:
@oldmoon_1 反原発論者で悪目立ちしてる人たちに関しては、「自分たちに同調しないものはみんな敵だ」という排他的な印象が確かに強いですね。「今は無理でも最終的には脱原発を」っていう人たちとの間にも明確な溝がありますし(この溝こそが彼らの最大の問題と個人的には思っています)。
原発推進と大見得を切って言えるはずなどないにしろ、原発依存の段階的脱却の議論に水を差したり、目くらましにモラルハザードが利用されてる現状は目を覆いたくなる。国策レベルに虚偽の嵐が影響を及ぼすべきではない。
□ 怒らないで聴いて欲しいんだけど、『有名人』で、フォロワーが一万人以上いる人が、「影響力が大きい」という自負から、他人と別段差異のある主張でもない当たり前のことを、さも啓示のごとく大衆に発信するのって、それって「義務感」でやらなければいけないことなんですかね。
□
TomoMachi:
自民党の憲法改正草案http://bit.ly/SODuHJ 第百二条「全て国民は、この憲法を尊重しなければならない」自民党内に「医師法が医師に対する規定であるように、憲法は国家に対する規定であって、国民に対するものではないですよ」と義務教育レベルの指摘をする人はいなかったのか…
為政者が「国民がどうこうあるべき」って語るのって片腹痛いというか、構造上の論理の無限後退に陥ってると気付けないのは、やはり無自覚に「傲り」に囚われるせいなのかとか
□
tokoroten:
普通選挙とは、統計学の成果を全否定しランダムサンプリングに納得できない人が納得するために全数検査を行い莫大なコストを支払う儀式である。 ってのが100年後くらいの教科書に載っていそう。 #人類が皆統計学を理解する日は来ない
ランダムサンプリングによる有権者調査で議席数が分かるなら、選挙権って要らないんじゃないかなぁ。 選挙速報にしても、開票率0%で当選確実が出るような世界だし。 政治家が統計を使わないことを馬鹿だというのであれば、統計の力によって議員を選出しないのもまた馬鹿なんじゃないのかなぁ。
選挙の意義ってのは、どうしたってダブルスタンダードなんだよねw 国民一人一人の意思を反映とか言って、結果は比率が全てで、少数派の反映などされない。民主主義は多数決であることと、一人一人が必ず投票しなければならない、この間に論理的飛躍があることに目を背けることが好都合。
意思決定の手続きとして選挙がある。ランダムサンプリングは、その結果を予測するのだから、選挙という経過を抜くと全体の動向にノイズが映る。とか