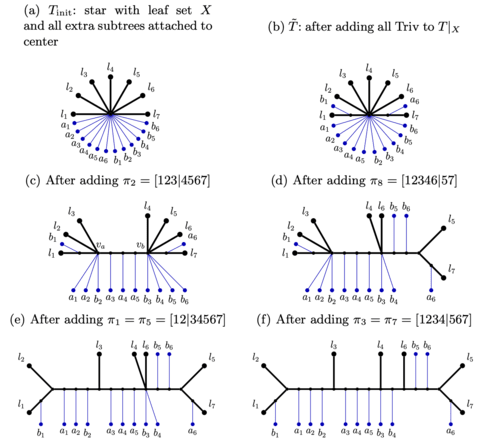
□ nanoSHAPE: Direct detection of RNA modifications and structure using single molecule nanopore sequencing
>>
https://www.biorxiv.org/content/10.1101/2020.05.31.126763v1.full.pdf
nanoSHAPE, a Direct RNA nanopore sequencing of AcIm modified RNA demonstrates significant promise to dissect complex RNA structures. Dissecting long-range structural elements that may orchestrate or result from alternative splicing may shed light on regulatory mechanisms underlying this complex phenomena.
□ On abstract F-systems. A graph-theoretic model for paradoxes involving a falsity predicate and its application to argumentation frameworks
>>
https://arxiv.org/abs/2005.07050
A Kripke's style fixed point characterization of groundedness is offered and fixed points which are complete (every sentence is deemed either true or false) and consistent (meaning that no sentence is deemed true and false) are put in correspondence with conglomerates.
the F-systems model abstracting from all the features of the language in which the represented sentences are expressed. the notion of conglomerate, the existence of which guarantees the absence of paradox.

□ Scale free topology as an effective feedback system
>>
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007825
A simplified approximation for complex structured networks which captures their dynamical properties. Constructing an approximation by lumping these nodes into a single effective hub, which acts as a feedback loop with the rest of the nodes.
a parametrization of scale free topology which is predictive at the ensemble level and also retains properties of individual realizations. the mean field theory predicts a transition between convergent and divergent dynamics which is corroborated by numerical simulations.
the role of outgoing hubs, in remarkable contrast to incoming ones, can be considered analogous to that of an external input in overcoming recurrent activity, suppressing chaotic dynamics, and ultimately driving the system to a stable fixed point.
The probability of convergence to a fixed point or QFP was calculated by simulating the dynamics of a an ensemble of 500 networks, and measuring the fraction of networks in the ensemble which reached the relevant frozen core criterion.

□ Network reconstruction for trans acting genetic loci using multi-omics data and prior information
>>
https://www.biorxiv.org/content/10.1101/2020.05.19.101592v1.full.pdf
a novel approach for understanding the molecular mechanisms underlying the statistical associations of trans -QTL hotspots by integrating existing biological knowledge and available multi-omics data to infer regulatory networks.
a comprehensive set of continuous priors from public datasets such as GTEx, the BioGrid and applied network inference incl glasso, BDgraph and iRafnet, that methods using data-driven priors outperform non-prior approaches for network reconstruction on simulated data.

□ Bio-semantic relation extraction with attention-based external knowledge reinforcement
>>
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-3540-8
a novel system that extends a Bidirectional Long Short-Term Memory neural network (BiLSTM) by taking advantage of knowledge from KBs.
Train the word embeddings, which include 1,701,632 vectors of distinct terms. They are represented by using word2vec based on 10,876,004 MEDLINE abstracts. Thus, each word is described as a 200-dimensional vector.

□ KOMB: Taxonomy-oblivious Characterization of Metagenome Dynamics via K-core Decomposition
>>
https://www.biorxiv.org/content/10.1101/2020.05.21.109587v1.full.pdf
K-core performs hierarchical decomposition which partitions the graph into shells containing nodes having degree at least K called K-shells, yielding O(E + V ) complexity.
the K-core of a graph is defined as the maximal induced subgraph where every node has (induced) degree at least K. Based on this sequence of K-cores, a node belongs to the K-shell if it is contained in the K-core but not in the (K+1)-core.
□ A2G2: A Python wrapper to perform very largealignments in semi-conserved regions
>>
https://www.biorxiv.org/content/10.1101/2020.05.21.109009v1.full.pdf
Amplicons to Global Gene is a Python wrapper that uses MAFFT and an “Amplicon to Gene” strategy to align very large numbers of sequences while improving alignment accuracy.
A2G2 uses the implementation of isolation forest available in Scikit-learn. Once the isolation forest method has identified entropic outliers, A2G2 will remove those query sequences from the alignment on request.
□ ARTDeco: automatic readthrough transcription detection
>>
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03551-
ARTDeco robustly quantifies the global severity of readthrough phenotypes, and reliably identifies individual genes that fail to terminate, are aberrantly transcribed due to upstream termination failure (read-in genes), and novel transcripts created as a result of readthrough.
ARTDeco can correct deconvolute the contribution of upstream readthrough transcription to total gene expression by using the upstream read-in expression.

□ DeepGraphMol: a multi-objective, computational strategy for generating molecules with desirable properties: a graph convolution and reinforcement learning approach
>>
https://www.biorxiv.org/content/10.1101/2020.05.25.114165v1.full.pdf
A molecule is generated by the Reinforcement Learning (RL) pathway using a Graph Convolutional Policy Networks. This molecule is then used as an input for the property prediction module which outputs the property score as predicted by the module.
Each intermediate layer consists of a Linear Layer followed by ReLU activation and Dropout that map the hidden vector to another vector of the same size. Finally the penultimate nodes are passed through a Linear Layer to output the predicted property score.
DeepGraphMol treats graph generation as a Markov Decision Process such that the next action is predicted based only on the current state of the molecule, not on the path that the generative process has taken.
□ mbkmeans: fast clustering for single cell data using mini-batch k-means
>>
https://www.biorxiv.org/content/10.1101/2020.05.27.119438v1.full.pdf
The mbkmeans follows a similar iterative approach to Lloyd’s algorithm. The mbkmeans software package implements the mini-batch k-means clustering algorithm described above and works with matrix-like objects as input.
The mbkmeans truly scalable and applicable to both standard in-memory matrix objects, including sparse matrix representations, and on-disk data representations that do not require all the data to be loaded into memory at any one time.
□ ENANO: Encoder for NANOpore FASTQ files
>>
https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btaa551/5848644
ENANO algorithm consistently achieves the best compression performance on every nanopore dataset, while being computationally efficient in terms of speed and memory requirements when compared to existing alternatives.
ENANO offers two modes, Maximum Compression and Fast (default), which trade-off compression efficiency and speed. in terms of encoding and decoding speeds, ENANO is 2.9x and 1.7x times faster than SPRING.

□ Two-pass alignment using machine-learning-filtered splice junctions increases the accuracy of intron detection in long-read RNA sequencing
>>
https://www.biorxiv.org/content/10.1101/2020.05.27.118679v1.full.pdf
To simulate basecall errors, sequences were inverted to the 3′ → 5′ direction and reads were generated using Markov chain Monte Carlo simulations with the basecall model.
The per-splice junction JAD was calculated as the maximum of the per-read JADs. Using the recommended alignment parameters for minimap but with the splicejunction filtering parameters as were used for nanopore DRS data.

□ BioSWITCH: Switching On Static Gene Regulatory Networks to Compute Cellular Decisions
>>
https://www.biorxiv.org/content/10.1101/2020.05.29.122200v1.full.pdf
BioSWITCH, a command-line program using the BioPAX standardised language to "switch on" static regulatory networks so that they can be executed in GINML to predict cellular behaviour.
BioSWITCH successfully and faithfully automates the network de-coding and re-coding into an executable logical network. BioSWITCH also supports the integration of a BioPAX model into an existing GINML graph.

□ TreeMap: A Structured Approach to Fine Mapping of eQTL Variants
>>
https://www.biorxiv.org/content/10.1101/2020.05.31.125880v1.full.pdf
TreeMap identifies putative causal variants in cis-eQTL accounting for multisite effects and genetic linkage at a locus. Guided by the hierarchical structure of linkage disequilibrium, TreeMap performs an organized search for individual and multiple causal variants.
TreeMap uses a nested model that first employs the tree-guided lasso algorithm to scan a large genomic region for candidate loci and candidate variants within a locus, and then apply statistical inference to derive credible sets of putative causal variants.
□ TransIntegrator: Integrate Heterogeneous NGS and TGS Data to Boost Genome-free Transcriptome Research
>>
https://www.biorxiv.org/content/10.1101/2020.05.27.117796v1.full.pdf
TransIntegrator generates a library of full expressed transcripts for an organism in a genome-free manner via integrating multiple heterogeneous sequencing datasets.
an example of using heterogeneous data maximally to overcome the gap of unsatisfied genome or no genome. TransIntegrator broadens the gene exploration from the static state to the dynamic state by providing a nearly complete transcript library as alternative reference to genome.

□ SuperFreq: Detecting copy number alterations in RNA-Seq
>>
https://www.biorxiv.org/content/10.1101/2020.05.31.126888v1.full.pdf
SuperFreq identifies the copy number alterations and point mutations in each clone, and highlights potentially causing mutations through variant annotation and COSMIC.
SuperFreq defines 𝑤i = 𝑙i/𝑡i, the scale factor of the limma t-distribution, as a measure of the variance of the LFC in the downstream segmentation of the genome.

□ scclusteval: Evaluating single-cell cluster stability using the Jaccard similarity index
>>
https://www.biorxiv.org/content/10.1101/2020.05.26.116640v1.full.pdf
scclusteval and the accompanying Snakemake workflow implement all steps of the pipeline: subsampling the cells, repeating the clustering with Seurat, and estimation of cluster stability using the Jaccard similarity index.
and for each cluster a Jaccard index is calculated to evaluate cluster similarity before and after re-clustering. Repeat the re-clustering for a number of times and use the mean or median of the jaccard indices as a metric to evaluate the stability of the cluster.
□ HAST: Haplotype-Resolved Assembly for Synthetic Long Reads Using a Trio-Binning Strategy
>>
https://www.biorxiv.org/content/10.1101/2020.06.01.126995v1.full.pdf
HAST exports two haplotypes of a diploid species for synthetic long reads with trio binning. HAST to recovers haplotypes with a scaffold N50 of >11 Mb and an assembly accuracy of 99.99995% (Q63).
HAST employs the haplotype-specific k-mers from parents to partition the SLR long fragments, and individually de novo assembles them to accurately construct the haplotypes with the long-range information.
□ MetaCNV - A Consensus Approach to Infer Accurate Copy Numbers From Low Coverage Data
>>
https://pubmed.ncbi.nlm.nih.gov/32487140/
MetaCNV integrates the results of multiple copy number callers and infers absolute and unbiased copy numbers for the entire genome.
MetaCNV is based on a meta-model that bypasses the weaknesses of current calling models while combining the strengths of existing approaches. MetaCNV is based on ReadDepth, SVDetect, and CNVnator.

□ scNym: Semi-supervised adversarial neural networks for single cell classification
>>
https://www.biorxiv.org/content/10.1101/2020.06.04.132324v1.full.pdf
scNym uses the unlabeled target data through a combination of MixMatch semi-supervision. The MixMatch semi-supervision combines MixUp data augmentations with pseudolabeling of the target data to improve generalization across the training and target domains.
scNym uses domain adversarial networks (DAN) as an additional approach to incorporate information from the target dataset. Entropy minimization and domain adversarial training enforce an inductive bias that all cells in the target dataset belong to a class.
□ GeneRax: A Tool for Species Tree-Aware Maximum Likelihood Based Gene Family Tree Inference Under Gene Duplication, Transfer, and Loss
>>
https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msaa141/5851843
GeneRax can infer rooted phylogenetic trees for multiple gene families, directly from the per-gene sequence alignments and a rooted, yet undated, species tree.
GeneRax does not require bootstrap-support thresholds, parsimony weights, MCMC convergence criteria, chain settings, proposal tuning, or priors. GeneRax infers trees that are the closest to the true tree in 90% of the simulations in terms of relative Robinson-Foulds distance.
□ MetaPhat: Detecting and Decomposing Multivariate Associations From Univariate Genome-Wide Association Statistics
>>
https://www.frontiersin.org/articles/10.3389/fgene.2020.00431/full
metaCCA extended CCA to work directly from GWAS summary statistics (effect size estimates and standard errors) of related traits and studies.
MetaPhat (Meta-Phenotype Association Tracer), a novel method to efficiently and systematically identify and annotate significant variants via multivariate GWAS from univariate summary statistics using metaCCA.
□ sPLINK: A Federated, Privacy-Preserving Tool as a Robust Alternative to Meta-Analysis in Genome-Wide Association Studies
>>
https://www.biorxiv.org/content/10.1101/2020.06.05.136382v1.full.pdf
sPLINK is robust against the heterogeneity of phenotype distributions across the cohorts. sPLINK always delivers the same p-values as aggregated analysis and correctly identifies all significant SNPs independent of the phenotype distributions across the separate datasets.
sPLINK implements a horizontal federate learning to preserve the privacy of data. sPLINK also provides a chunking capability to handle large datasets containing millions of SNPs, and supports multiple association tests incl. chi-square, linear regression, and logistic regression.

□ Matrix factorization with neural network for predicting circRNA-RBP interactions https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-3514-x
Usually, the architecture of neural network has a significant impact on its performance, especially the depth of network is a prominent impact factor.
Matrix factorization based on neural network kernel model, a computational framework to predict unknown circRNA-RBP interaction pairs with Positive-Unlabeled (P-U) learning.

□ Low Dimensionality in Gene Expression Data Enables the Accurate Extraction of Transcriptional Programs from Shallow Sequencing
>>
https://www.cell.com/cell-systems/fulltext/S2405-4712(16)30109-0
a mathematical framework that defines this tradeoff between mRNA-sequencing depth and error in the extraction of biological information. Transcriptional programs can be reproducibly identified at 1% of conventional read depths.
a simple read depth calculator that determines optimal experimental parameters to achieve a desired analytical accuracy. shallow mRNA-seq is similarly enabled by an inherent low dimensionality in gene expression datasets that emerges from groups of covarying genes.

□ scASK: A novel ensemble framework for classifying cell types based on scRNA-seq data
>>
https://www.biorxiv.org/content/10.1101/2020.06.07.138271v1.full.pdf
Adaptive Slice KNNs (scASK) onsists of three innovational modules, called DAS (Data Adaptive Slicing), MCS (Meta Classifiers Selecting) and EMS (Ensemble Mode Switching), respectively, which facilitate scASK to approximate a bias-variance tradeoff beyond classification.
scASK is the first generic ensemble classification framework especially for classifying cell types based on scRNA-seq data with high dimensionality. scASK applies the known cell-type labels to new samples with high accuracy and high robustness in a near-real time.
□ IGD: high-performance search for large-scale genomic interval datasets
>>
https://www.biorxiv.org/content/10.1101/2020.06.08.139758v1.full.pdf
integrated genome database, a method and tool for searching genome interval datasets more than three orders of magnitude faster than existing approaches, while using only one hundredth of the memory.
IGD uses a novel linear binning method that allows us to scale analysis to billions of genomic regions. the linear binning is a single-level binning. The whole genome is divided into equal-sized bins and intervals are put into bins they intersect.

□ Monet: An open-source Python package for analyzing and integrating scRNA-Seq data using PCA-based latent spaces
>>
https://www.biorxiv.org/content/10.1101/2020.06.08.140673v1.full.pdf
At its core, Monet implements algorithms to infer the dimensionality and construct a PCA-based latent space from a given dataset, with the dimensionality being automatically determined using molecular cross-validation.
This latent space, represented by a MonetModel object, then forms the basis for data analysis and integration. Monet’s MCV-based inference provides a much more systematic way of determining the dimensionality.
□ Higher order Markov models for metagenomic sequence classification
>>
https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btaa562/5855128
A novel software implementation performs classification faster than the existing Markovian metagenomic classifiers and can therefore be used as a standalone classifier or in conjunction with existing taxonomic classifiers for more robust classification of metagenomic sequences.
Single-Order Markov Model Builder reports the log-probability that a read originated from a genome based on the single-order markov model constructed from all of the information contained within that genome.
□ ATAV - a comprehensive platform for population-scale genomic analyses
>>
https://www.biorxiv.org/content/10.1101/2020.06.08.136507v1.full.pdf
the ATAV framework allows continuous real time analyses of all samples loaded into the database without the need for computationally demanding joint calling preceding each analysis and it allows convenient tracking of precise analyses performed.
ATAV stores variant and coverage data for all samples in a centralized database, which is then efficiently queried by ATAV to support diagnostic analyses for trios and singletons, as well as rare-variant collapsing analyses for finding disease associations in complex diseases.
□ Spritz: A Proteogenomic Database Engine
>>
https://www.biorxiv.org/content/10.1101/2020.06.08.140681v1.full.pdf
Spritz automatically sets up and executes approximately 20 tools, which enable construction of a proteogenomic database from only raw RNA sequencing data.
The combination of a Spritz database and the use of G-PTM-D within MetaMorpheus allows for identification of post-translationally modified variant. no other search strategies are capable of detecting post-translationally modified variant sites.

□ Converting networks to predictive logic models from perturbation signalling data with CellNOpt
>>
https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btaa561/5855133
an efficient Integer Linear Programming (ILP), a probabilistic logic implementation for semi-quantitative datasets, the integration of a stochastic Boolean simulator, a tool to identify missing links, systematic post-hoc analyses, and an R-Shiny tool to run CellNOpt interactively.
Dynamic-Feeder is a method to identify missing links in the PKN and provides candidates to fill knowledge gaps. Dynamic-Feeder generalizes the CNORfeeder to time-course data with a logic ordinary differential equations formalism.

□ FASTQuick: Rapid and comprehensive quality assessment of raw sequence reads
>>
https://www.biorxiv.org/content/10.1101/2020.06.10.143768v1.full.pdf
FASTQuick offer orders of magnitude faster turnaround time than existing full alignment-based methods while providing comprehensive and sophisticated quality metrics, including estimates of genetic ancestry and contamination.
By focusing on a variant-centric subset of a reference genome(reduced reference genome), FASTQuick offer up to 30~100-fold faster turnaround time than existing post-alignment methods for deeply sequenced genome.
□ BoardION: real-time monitoring of Oxford Nanopore Technologies devices
>>
https://www.biorxiv.org/content/10.1101/2020.06.09.142273v1.full.pdf
BoardION’s dynamic and interactive interface allows users to explore sequencing metrics easily and quickly and to optimize in real time the quantity and the quality of the generated data.
the existing ONT interface (MinKnow) does not provide enough plots or interactivity to explore the quality of sequencing data in depth. BoardION dedicated to sequencing platforms, for real-time monitoring of all ONT sequencing devices; MinION, Mk1C, GridION and PromethION.
□ Amino acid encoding for deep learning applications
>>
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03546-x
In deep learning applications, discrete data, e.g. words or n-grams in language, or amino acids or nucleotides in bioinformatics, are generally represented as a continuous vector through an embedding matrix.
End-to-end learning is a flexible and powerful method for amino acid encoding. Its schemes should be benchmarked against random vectors of the same dimension to disentangle the information content provided by the encoding scheme from the distinguishability effect.

□ Trans-NanoSim Characterizes and Simulates Nanopore RNA-sequencing Data
>>
https://academic.oup.com/gigascience/article/9/6/giaa061/5855462
Trans-NanoSim is the first transcriptome sequence simulator that provides IR modelling. Considering the human direct RNA dataset as an example.
the IR modelling module of Trans-NanoSim identified 2,872 transcripts with ≥1 retained intron, and nearly half of them (1,285 transcripts) were expressed at >2 transcripts per million.
□ Fast and Accurate Prediction of Partial Charges Using Atom-Path-Descriptor-based Machine Learning
>>
https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btaa566/5856101
In the APD algorithm, the 3D structures of molecules were assigned with atom centers and atom-pair path-based atom layers to characterize the local chemical environments of atoms.
Atom Path Descriptor empolys random forest (RF) and extreme gradient boosting (XGBoost) to develop the regression models for partial charge assignment.
□ GAD: A Python Script for Dividing Genome Annotation Files into Feature-Based Files
>>
https://link.springer.com/article/10.1007/s12539-020-00378-4
GAD is a cross-platform graphical interface tool used to extract genome features such as intergenic regions, upstream, and downstream genes.
GAD finds all names of ambiguous sequence ontology, and either extracts them or considers them as genes or transcripts. The GAD can handle large sizes of different genomes and an infinite number of files.

□ ATS: Detecting differential alternative splicing events in scRNA-seq with or without Unique Molecular Identifiers
>>
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007925
SCATS identified more differential splicing events with subtle difference across cell types compared to Census and DEXSeq.