
□ London Calling 2019: Nanopore Conference #NanoporeConf
>> https://londoncallingconf.co.uk/lc19
>> https://nanoporetech.com
Plenary and breakout presentations on the latest research using nanopore sequencing, live product demonstrations, practical clinics, evening networking events and much more.
□ H.E.L.E.N. (Haplotype Embedded Long-read Error-corrector for Nanopore):
>> https://github.com/kishwarshafin/helen
HELEN is a polisher intended to use for polishing human-genome assemblies generated by the Shasta assembler.
HELEN uses a Recurrent-Neural-Network (RNN) based Multi-Task Learning (MTL) model that can predict a base and a run-length for each genomic position using the weights generated by MarginPolish.
MarginPolish uses a probabilistic graphical-model to encode read alignments through a draft assembly to find the maximum-likelihood consensus sequence. The graphical-model operates in run-length space, which helps to reduce errors in homopolymeric regions.
□ Shasta long read assembler:
>> https://chanzuckerberg.github.io/shasta/
The goal of the Shasta long read assembler is to rapidly produce accurate assembled sequence using as input DNA reads generated by Oxford Nanopore flow cells.
Using a run-length representation of the read sequence. This makes the assembly process more resilient to errors in homopolymer repeat counts, which are the most common type of errors in Oxford Nanopore reads.
□ Nanopore's Long DNA Paradox:
>> https://omicsomics.blogspot.com/2019/05/nanopores-long-dna-paradox.html
How does DNA choke a pore? Why does ultra-long DNA seem to be worse? These are mysteries.
front-and-center in the plant genomics sub-session I attended and could be called the central paradox of the current state of nanopore sequencing: pores are great for long DNA but long DNA is not great for pores.


□ OmicsOmicsBlog: #NanoporeConf different complexities of repeats.
□ gringene_bio: #NanoporeConf predictions; maybe, maybe not (they'll happen when they happen):
* 1000 bases / second [slowly getting ducks in a row]
* Solid state
* VolTRAX / MinION hybrid (TraxION)
* SmidgION
* Ubik tube
Based mostly on Clive's last NCM talk, here are my #NanoporeConf tech update predictions, starting with an almost-certain accuracy update:
* R10 everywhere
* base caller / polishing improvements
* mumbling about homopolymers
* magic 8-base PCR mix
* Linear consensus

□ libarbaraa: This map shows where MinION has been used. And they've just announced that they are willing to expand MinION usage in Africa.
□ Revealing mRNA alternative splicing complexity in the human brain':
https://vimeo.com/337887055
□ NanoporeConf: Michael Boemo of Oxford presenting on the ability of ultra-long Nanopore reads to map DNA replication dynamics, including the detection of these origins within repetitive regions and in cis to enable the study of multiple origins along a single molecule. #Nanoporeconf

□ RNAkook: Christopher Oakes bringing complex EBV methylation patterns to light using #nanopore sequencing #NanoporeConf
□ marimiya_tky: NanoGalxy is available here! https://nanopore.usegalaxy.eu/

□ DrT1973: It’s not just for DNA/RNA, great to see‘s talk at #nanoporeconf on protein detection with the nanopore “molecular scale” sensing device.
□ Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen
>> https://www.nature.com/articles/s41467-019-08734-9
direct RNA-seq to profile the herpes simplex virus type 1 (HSV-1) transcriptome during productive infection of primary cells. direct RNA-seq offers a powerful method to characterize the changing transcriptional landscape of viruses with complex genomes.
□ UNCALLED: A Utility for Nanopore Current Alignment to Large Expanses of DNA
>> https://github.com/skovaka/UNCALLED
read-until with UNCALLED - stepwise behavior due to API limitation.

□ UNCALLED: #NanoporeConf Sam Kovac JHU Read until Matt Loose method only up to 10kob hence UNCALLED maps raw signal to 10s of megabases using knees and allpaths using FM index that scales with query, not genome

□ NanoporeConf: Michael Boemo of Oxford presenting on the ability of ultra-long Nanopore reads to map DNA replication dynamics, including the detection of these origins within repetitive regions and in cis to enable the study of multiple origins along a single molecule. #Nanoporeconf
□ ppamaral: @tom_leon
introducing Nanocompore to detect different RNA modifications in dRNA-seq using Nanopore.

□ Transient crosslinking kinetics optimize gene cluster interactions
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/23/648196.full.pdf
computational modeling of the full genome during G1 in budding yeast, exploring four decades of timescales for transient crosslinks between 5kbp domains (genes) in the nucleolus on Chromosome XII;
temporal network models with automated community (cluster) detection algorithms applied to the full range of 4D modeling datasets.
"rigid" clustering emerges with clusters that interact infrequently; with longer crosslink lifetimes, there is a dissolution of clusters.
□ Nanopore sequencing in space: one small step for MinION, one giant leap for spaceflight research
Sanger sequencing confirmation of species level IDs from extraterrestrial sequencing on the ISS
□ Cyclomics: ultra-sensitive detection of cell-free tumour cfDNS
>> https://www.lifesciencesatwork.nl/profile/cyclomics/
Mutation detection in signal space using Dynamic Time Warping.
improvements to accuracy with guppy high-accuracy basecaller when identifying TP53 mutation.
□ in Africa, Charles Kayuki used MinION + PDQex to minimise sequencing effort. A 2hr run off battery with MinIT worked for identification. Power is an issue; doesn't recall a 48hr run that could finish before the power droppedout.

□ ChiCMaxima: a robust and simple pipeline for detection and visualization of chromatin looping in Capture Hi-C
>> https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1706-3
ChiCMaxima, which uses local maxima combined with limited filtering to detect DNA looping interactions, integrating information from biological replicates.
ChiCMaxima gave a higher enrichment for interactions containing hallmarks of regulatory chromatin, such as histone modifications indicative of enhancers or CTCF binding sites, suggesting that its false positive detection rate for functional chromatin loops.

□ Direct RNA with @nanopore will get closer to mainstream method this year
on cDNA upgrades. 200 millions reads per promethion flowcell.
with cDNA improvements, you can now expect ~20 million reads from a MinION flow cell and 100 million reads from a PromethION flow cell (assuming 1kb transcript length)

□ OmicsOmicsBlog: Heron: improving single molecule accuracy. 1D^2 will be supported but not developed; “many horses in the race”.
RCA. Only get template strand - but lack of reannealing avoids signal shifting - but don’t have orthogonal data.

□ Daniela Bezdan: Nanopore include #UMI,#rollingcircle , circular and linear

□ raw 1D basecalling. Major algorithm improvements have been delivered ~annually: from HMM, to RNN events, to RNN transducer, to RNN on raw signal, and now flip-flop.
□ Plongle is essentially a 96 well plate compatible Flongle, targeting $25-$50 per well and we aim to have it out next year.

□ normal sample and right side cancer sample. SV landscape is totally difference. Some day we will use the circle diagram to predict, just like we used to use FISH

□ Molecular tagging with nanopore-orthogonal DNA strands
□ The first Run of P48@GrandOmics is 4.88Tb/42Cell in 96 hours.

□ Irina was somewhat successful with in-vitro tRNA, with polyA tailing and local alignment, but had trouble with total native tRNA due to modifications (30 modifications per 70 nucleotides). Will be trying custom base calling in the future.

□ COBS: a Compact Bit-Sliced Signature Index
>> https://arxiv.org/pdf/1905.09624.pdf
COBS, a compact bit-sliced signature index, which is a cross-over between an inverted index and Bloom filters.
the target application is to index k-mers of DNA samples or q-grams from text documents and process approximate pattern matching queries on the corpus with a user- chosen coverage threshold.
Query results may contain a number of false pos- itives which decreases exponentially with the query length and the false pos- itive rate of the index determined at construction time.
□ 10x single cell protocol has opportunities to adapt for long reads -
@ClarksysCorner
chooses to split GEMs, allowing selection of numbers of cells and depth of coverage for different sequencing purposes.

□ Comparing read numbers and QC info for different platforms reveals a few things. PromethION can give high read depth, making the depth of coverage per cell equivalent to short reads.

□ Mark Ebbert Dark by depth or dark by MapQ regions of the genome.

□ Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight
>> https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1707-2
identify regions with few mappable reads that we call dark by depth, and others that have ambiguous alignment, called camouflaged.
Linked-read or long-read sequencing technologies from 10x Genomics, PacBio, and Oxford Nanopore Technologies reduce dark protein-coding regions to approximately 50.5%, 35.6%, and 9.6%, respectively.

□ a comparison of R10 and R9.4 data, both native and PCR. We can get better than Q40 genomes on nanopore
R10. Better yet, our data is open, and can be downloaded right now: https://lomanlab.github.io/mockcommunity/r10.html

□ Visualising “the whale”: a 2.3Mb read from NH
@DeepSeqNotts
with MinION, a portable affordable native nucleus acids sensor #NanoporeConf
□ Enabling high-accuracy long-read amplicon sequences using unique molecular identifiers and Nanopore sequencing
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/645903.full.pdf
Partitioning based methods such as 10x Genomics and TruSeq Synthetic Long- Reads struggle resolve complex amplicon populations, as there is a high risk of >1 amplicon ending up in the same partition which will result in a chimeric assembly.
a UMI design containing recognizable internal patterns, which together with UMI length filtering now makes it possible to robustly determine true UMI sequences in raw nanopore data.
□ EnImpute: imputing dropout events in single cell RNA sequencing data via ensemble learning
>> https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btz435/5498284
EnImpute combines the results obtained from multiple imputation methods to generate a more accurate result.
The EnImpute package has the following R-package dependencies: DrImpute, Rmagic, rsvd, SAVER, Seurat, scImpute, scRMD and stats. The dependencies will be automatically installed along with EnImpute.

□ TeXP: Deconvolving the effects of pervasive and autonomous transcription of transposable elements
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/648667.full.pdf
TeXP builds mappability signatures from LINE-1 subfamilies to deconvolve the effect of pervasive transcription from autonomous LINE-1 activity.
validated TeXP by independently estimating the levels of LINE-1 autonomous transcription using ddPCR, finding high concordance.

□ Algorithms for efficiently collapsing reads with Unique Molecular Identifiers
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/648683.full.pdf
formulate the problem as a dynamic query problem that involves a common interface, and show that previous deduplication algorithms can be implemented with this interface no change to their results.
multiple data structures that implements this interface, and find that the n-grams BK-trees data structure is the most efficient through an empirical evaluation with simulated datasets.
If a significant portion of the UMI sequences share the same n-gram, then the algorithms will run as fast as just using one large BK-tree for all UMI sequences, which takes O(kR + k log N ) time, not O(N ) time.


□ Bayesian Item Response Modelling in R with brms and Stan
>> https://arxiv.org/pdf/1905.09501.pdf
how to use the R package brms together with the probabilistic programming language Stan to specify and fit a wide range of Bayesian IRT models using flexible and intuitive multilevel formula syntax.
For increased efficiency, defining both gamma and logitgamma as non-linear parameters and related them via gamma ~ inv_logit(logitgamma).
□ Innovative strategies for annotating the “relationSNP” between variants and molecular phenotypes
>> https://biodatamining.biomedcentral.com/articles/10.1186/s13040-019-0197-9
Synonymous variants are often grouped as one type of variant, however there are in fact many tools available to dissect their effects on gene expression.
ENCODE and GTEx have made it possible to annotate non-coding regions. Although annotating variants is a common technique among human geneticists, the constant advances in tools and biology surrounding SNPs requires an updated summary of what is known and the trajectory of the field.
□ Information Theoretic Feature Selection Methods for Single Cell RNA-Sequencing
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/646919.full.pdf
Unlike differential methods, that are strictly binary and univariate, information-theoretic methods can be used as any combination of binary or multiclass and univariate or multivariate.
A fast computation of entropy for sparse matrices. The time complexity of EntropyWRT is O(n + mp + q + mkr+1) where p is the number of rows with non-zero entries in columns j1, j2, ..., jr, and q is the number of non-zero entries in M.

□ epiScanpy: integrated single-cell epigenomic analysis
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/648097.full.pdf
EpiScanpy enables preprocessing of epigenomics data as well as downstream analyses such as clustering, manifold learning, visualization and lineage estimation.
EpiScanpy allows for comparative analyses between -omics layers, and can serve as a framework for future single-cell multi-omics data integration. comparing multiple feature space constructions for epigenetic data and show the feasibility of common clustering, dimension reduction and trajectory learning techniques.

□ DeepGRN: Interpretable attention model in transcription factor binding site prediction with deep neural networks
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/648691.full.pdf
DeepGRN incorporates the attention mechanism with the CNNs-RNNs based model by applying attention normalization before or after the LSTM layer.
Convolutional and BiLSTM layers use both forward and reverse complement features as inputs. Attention weights are computed from hidden outputs of LSTM and then are used to compute the weighted representation Z through a Kronecker product. Z is flattened and fused with non-sequential features.
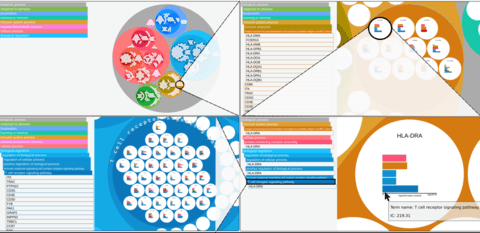
□ GSAn: an alternative to enrichment analysis for annotating gene sets
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/648444.full.pdf
The main problems in finding gene signatures are mainly related to the investigation of the biological function of gene sets. That problem can be solved using classical enrichment methods, such as DAVID or g:Profiler.
□ Genesis and Gappa: Library and Toolkit for Working with Phylogenetic (Placement) Data
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/24/647958.full.pdf
GENESIS is a highly flexible library for reading, manipulating, and evaluating phylogenetic data, and in particular phylogenetic placement data.
Gappa is a command line interface for analysis methods and common tasks related to phylogenetic placements. GSAn, a novel gene set annotation Web server that uses semantic similarity measures to reduce a priori Gene Ontology annotation terms.
□ MULKSG: MULtiple KSimultaneous Graph Assembly
>> https://link.springer.com/chapter/10.1007/978-3-030-18174-1_9
how to parallelize multi K de Bruijn graph genome assembly simultaneously, removing the bottleneck of iterative multi K assembly. a parallel version of the assembly and show the statistics are the same as when run on a single node.
The expected execution time on a single node with 40 cores is variable, with the average execution time for the entire pipeline over 16 datasets tested being 1613 s for SPAdes vs. 1581 s for MULKSG, with the MULKSG graph creation and traversal averaging 15% faster than SPAdes.
This algorithmic change gets rid of the single node sequential bottleneck on multi K genome assembly, allowing for the use of parallel error correction, graph building, graph correction, and graph traversal.
□ SELVa: Simulator of Evolution with Landscape Variation
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/25/647834.full.pdf
SELVa, the Simulator of Evolution with Landscape Variation, aimed at modeling the substitution process under a changing single position fitness landscape in a set of evolving lineages forming a phylogeny of arbitrary shape.
SELVa generates the root state for each position by sampling from the stationary distribution corresponding to the initial fitness vector.

□ Uncovering the structure of self-regulation through data-driven ontology discovery
>> https://www.nature.com/articles/s41467-019-10301-1
□ Comprehensive Multiple eQTL Detection and Its Application to GWAS Interpretation
>> https://www.genetics.org/content/early/2019/05/22/genetics.119.302091
a statistical pipeline to achieve the following goals: (a) to evaluate the prevalence of multiple cis-eQTL regulation in human peripheral blood; (b) to estimate the extent of QTL signal sharing across three expression platform;
and (c) to detect co-localization of eQTL signals with GWAS hits contingent on the LD at each locus, revealing the possible biological regulatory mechanisms linking genetic variants to complex human phenotypes.
□ HiChIRP reveals RNA-associated chromosome conformation
>> https://www.nature.com/articles/s41592-019-0407-x
HiChIRP, a method leveraging bio-orthogonal chemistry and optimized chromosome conformation capture conditions, which enables interrogation of chromatin architecture focused around a specific RNA of interest down to approximately ten copies per cell.
HiChIRP of three nuclear RNAs reveals insights into promoter interactions (7SK), telomere biology (telomerase RNA component) and inflammatory gene regulation (lincRNA-EPS).
□ Spectral clustering in regression-based biological networks
>> https://www.biorxiv.org/content/biorxiv/early/2019/05/27/651950.full.pdf
the effects of using estimates from regression models when applying the spectral clustering approach to community detection. We demonstrate the impacts on the affinity matrix and consider adjusted estimates of the affinity matrix for use in spectral clustering.
a recommendation for selection of the tuning parameter in spectral clustering. evaluate the proposed adjusted method for performing spectral clustering to detect gene clusters in eQTL data from the GTEx project and to assess the stability of communities in biological data.

□ snakePipes: facilitating flexible, scalable and integrative epigenomic analysis
>> https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz436/5499080
snakePipes, a workflow package for processing and downstream analysis of data from common epigenomic assays: ChIP-seq, RNA-seq, Bisulfite-seq, ATAC-seq, Hi-C and single-cell RNA-seq.
unlike conventional pipelines, workflows in snakePipes are based on a repository of modular rules, such that multiple variations of each workflow can be assembled on-the-fly by changing the parameters on their command-line wrappers.

□ Benchmarking of 4C-seq pipelines based on real and simulated data:
>> https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz426/5499078
a benchmarking study on 66 4C-seq samples from 20 datasets, and developed a novel 4C-seq simulation software, Basic4CSim, to allow for detailed comparisons of 4C-seq algorithms on 50 simulated datasets with 10 to 120 samples each.
For near-cis scenarios, r3Cseq, peakC, and FourCSeq offered high precision, while fourSig demonstrated high overall F1 scores in far-cis analyses.
□ clustermq enables efficient parallelisation of genomic analyses
>> https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz284/5499081
it hinders load balancing between computing nodes (as it requires a file- system based lock mechanism) and the use of remote compute facilities without shared storage systems.
clustermq distributes data over the network without involvement of network-mounted storage, monitors the progress of up to 10^9 function evaluations, and collects back the results.

□ ExpansionHunter: A sequence-graph based tool to analyze variation in short tandem repeat regions
>> https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz431/5499079
Expansion Hunter aims to estimate sizes of such repeats by performing a targeted search through a BAM/CRAM file for reads that span, flank, and are fully contained in each repeat.
ExpansionHunter translates each regular expression into a sequence graph. Informally, a sequence graph consists of nodes that correspond to sequences and directed edges that define how these sequences can be connected together to assemble different alleles.
a novel method that addresses the need for more accurate genotyping of complex loci. This method can genotype polyalanine repeats and resolve difficult regions containing repeats in close proximity to small variants and other repeats.
□ Deep Fusion of Contextual and Object-based Representations for Delineation of Multiple Nuclear Phenotypes
>> https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btz430/5499132
This Application-Note couples contextual information about the cellular organization with the individual signature of nuclei to improve performance. Routine delineation of nuclei in H&E stained histology sections is enabled for either computer-aided pathology or integration with genome-wide molecular data.
□ Nextpolish
>> https://github.com/Nextomics/NextPolish
NextPolish is used to fix base errors (SNP/Indel) in the genome generated by noisy long reads, it can be used with short read data only or long read data only or a combination of both.





















※コメント投稿者のブログIDはブログ作成者のみに通知されます