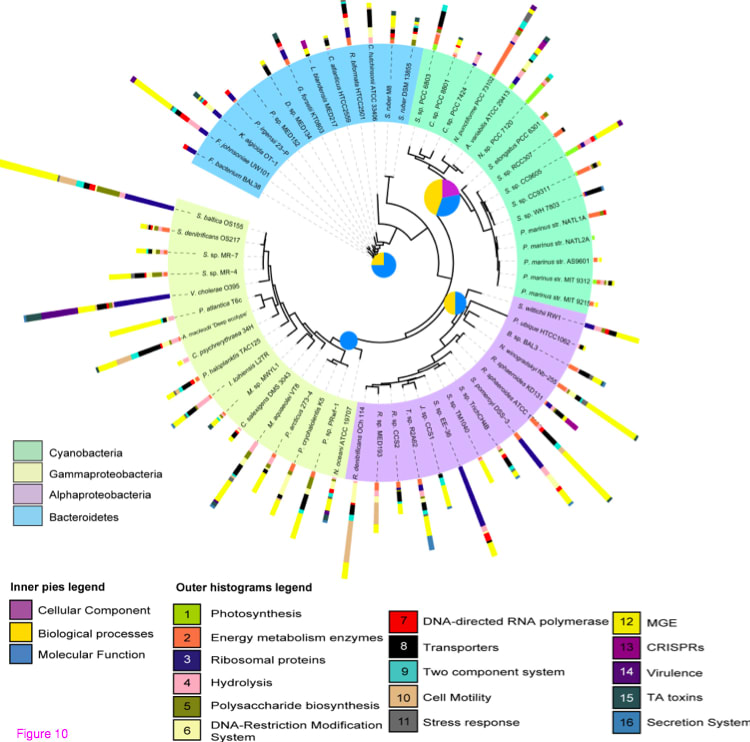
(Biologically relevant genes within marine bacterial GIs:16 biological categories with the potential to increase fitness)
□ Patterns and architecture of genomic islands in marine bacteria
>> http://www.biomedcentral.com/content/pdf/1471-2164-13-347.pdf
70 selected marine bacterial genomes detected with IslandViewer, to explore the distribution, patterns and functional gene content in these genomic regions. We detected 438 GIs containing a total of 8152 genes. We showed that the GI number per genome was strongly and positively correlated with the total GI size. In 50% of the genomes analyzed the GIs accounted for approximately 3% of the genome length, with a maximum of 12%.

□ Assessing Drug Target Association Using Semantic Linked Data
>> http://t.co/iV8EiVRv
a statistical model to assess the association of drug target pairs based on their relation with other linked objects. Validation experiments demonstrate the model can correctly identify known direct drug target pairs with high precision. Indirect drug target pairs (for example drugs which change gene expression level) are also identified but not as strongly as direct pairs. We further calculated the association scores for 157 drugs from 10 disease areas against 1683 human targets, and measured their similarity using a score matrix.

□ Bina is bringing an Apple-like business model to the rapidly expanding world of genomics.
>> http://tech.fortune.cnn.com/2012/07/11/remaking-genome-analysis/
The Bina Box is an Apple-like (AAPL) combination of company-developed hardware and software that work together to crunch sequenced genetic data faster than conventional solutions -- sometimes over 100 times faster in some cases.
The box analyzes and compresses those 300 gigabytes of raw data into several hundred megabytes of information -- making it much faster to upload -- and either transmits it to the Bina Cloud, which stores and processes the data, or a private cloud constructed for the company. That also allows users such as doctors and medical specialists to consume and share it. As a result, what once took several days to over a month to accomplish can take as little as two hours.


□ GenomeRing: alignment visualization based on SuperGenome coordinates
>> http://bioinformatics.oxfordjournals.org/content/28/12/i7.full
>> http://it.inf.uni-tuebingen.de/software/genomering/
First, we propose the SuperGenome concept for the computation of a common coordinate system for all genomes in a multiple alignment. This coordinate system allows for the consistent placement of genome annotations in the presence of insertions, deletions and rearrangements.多重アラインメントされたゲノムの共通座標系を用いた可視化ツール。Maydayプラットフォームに統合
Second, we present the GenomeRing visualization that, based on the SuperGenome, creates an interactive overview visualization of the multiple genome alignment in a circular layout.
now integrated the GenomeRing visualization with MAYDAY (Battke et al., 2010), our visual analysis platform for ‘omics’ data. As a result, GenomeRing can be linked with all other visualizations offered by MAYDAY, including a traditional, linear genome browser.

□ opentreeoflife:
Tree history RT @SpeechNerdess @brainpicker Visual history of evolution in tree-like diagrams http://j.mp/K9b1KH #boidiversity #treeoflife
Mapping 450 years of mankind’s curiosity about the living world and the relationships between organisms.
Origin of Species by Means of Natural Selection in 1859, the concept, most recently appropriated in mapping systems and knowledge networks, is actually much older, predating the theory of evolution itself. The collection is thus at once a visual record of the evolution of science and of its opposite ? the earliest examples, dating as far back as the sixteenth century, portray the mythic order in which God created Earth, and the diagrams’ development over the centuries is as much a progression of science as it is of culture, society, and paradigm.生物種ツリー・ダイアグラムの歴史。図象隠喩としては最も普遍的で、古くは12世紀ルルスのArs Magnaに遡る。シャフナーからポピュラーに。
M Gumbel - SCABIO: a framework for bioinformatics algorithms in Scala
View more PowerPoint from Jan Aerts
□ SCABIO: a framework for bioinformatics algorithms in Scala
>> http://www.mi.hs-mannheim.de/gumbel/en/forschung/scabio/
generic and extensible dynamic programming (DP) algorithm. Many bioinformatics algorithms rely on this method. scabio simply comes with a dynamic programming framework/interface which can be (re-)used or extended for many purposes, especially for all kind of bioinformatics algorithms. Within SCABIO, all pairwise alignment algorithms, the Viterbi-algorithm and the Nussinov-algorithm are based on these DP classes.先ほどのBOSCでアナウンスされた、バイオインフォマティクスScalaフレームワーク。僅かなコードベースで再利用性向上

□ Computational analysis of target hub gene repression regulated by multiple and cooperative miRNAs
>> http://nar.oxfordjournals.org/content/early/2012/07/13/nar.gks657.full
We then show how a kinetic model can be derived from the regulatory map. To validate our approach, we present a case study for p21, one of the first experimentally proved miRNA target hubs. Our analysis indicates that distinctive expression patterns for miRNAs, some of which interact cooperatively, fine-tune the features of transient and long-term regulation of target genes.数学的モデリングで生物医学と定量データを統合
With respect to p21, our model successfully predicts its protein levels for nine different cellular functions. In addition, we find that high abundance of miRNAs, in combination with cooperativity, can enhance noise buffering for the transcription of target hubs.
□ Zooma 2: A million gene expression annotations: Gene Expression Atlas, aligned to ontology classes in EFO.
>> http://wwwdev.ebi.ac.uk/fgpt/zooma/
>> http://t.co/T0wwjzjq
Linked data repos of annotation knowledge.
A million gene expression annotations with Zooma.
Zooma is an RDF knowledge base of annotation knowledge, extracted from the expert curation performed on a subset of ArrayExpress data. This subset of data has the added advantage of being curated twice because it has also been loaded into the Gene Expression Atlas, where it has been aligned to ontology classes in EFO. This is very powerful for several reasons.
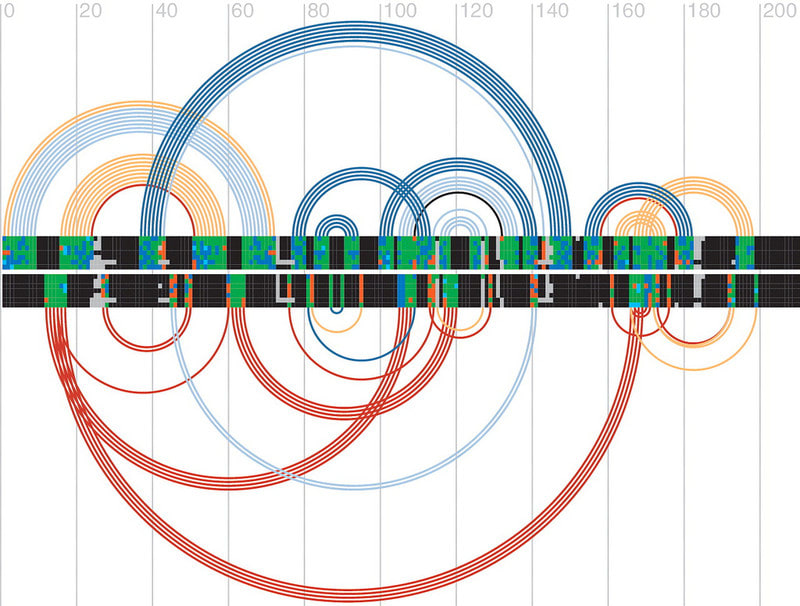
□ R-chie overlapping covariance arc diagram:
>> http://www.flickr.com/photos/dullhunk/7554750410/
RNA構造予測アルゴリズムを評価する為のアークダイアグラム。
□ A new entropy model for RNA: A critique of the standard Jacobson-Stockmayer model applied to multiple cross links
>> http://www.pagepress.org/journals/index.php/jnai/article/view/2650 …
CLEモデルにおける全球規模のエントロピー損失と計算粒度の指標
□ Evolutionary dynamics of RNA-like replicators: A bioinformatic approach to the origin of life:
>> http://www.narcis.nl/publication/RecordID/oai%3Adspace.library.uu.nl%3A1874%2F42375/uquery/RNA+world+and+RNA+world/id/2/Language/EN
□ Evolution of Protein Synthesis from an RNA World:
>> http://cshperspectives.cshlp.org/content/4/4/a003681.long …
This cycle of noncovalent complex formation, followed by ligation, can then be repeated, while selecting for improvement of ribosome-like function. Finally, the influence of the peptides produced by a primitive ribosome on its own structure and assembly would also begin to impact its own evolution. Ribosome-binding peptides may thus have played an early role in shaping the ultimate form (and function) of the ribosome.
リボソームのコーディング機構。非コードペプチド合成のテンプレート指向性システム。
□ assemblathon:
RNA-seq analysis of C. briggsae genome reveals >10,000 small assembly errors. Genome Research http://j.mp/OqbMMh
□ Smart-Seq: a robust mRNA-Seq protocol: Full-length Seq from single-cell levels of RNA individual circulating tumor cell nature.com/nbt/journal/va…
□ Genomicswatch:
Genomic sequencing method offers “smarter” cell analysis - R & D Magazine http://bit.ly/LHOtLZ
□ biosharing:
BioSharing and the ISA Commons presented at ?#ISMB? 2012; BOSC (P. Rocca-Serra) and at Highlight Track (S. Sansone): http://www.nature.com/ng/journal/v44 …

□ A graph-based approach for designing extensible pipelines
>> http://www.biomedcentral.com/content/pdf/1471-2105-13-163.pdf
a graph-based approach to implement extensible and low-maintenance pipelines that is suitable for pipeline applications with multiple functionalities that require different combinations of steps in each execution. Here pipelines are composed automatically by compiling a specialised set of tools on demand, depending on the functionality required, instead of specifying every sequence of tools in advance.Bioinformaticsパイプライン拡張性のグラフベース管理。発現解析など、複数ソフトウェアを並行する分野に応用可。
2011年頃から、Bioinformatics分野においても"Graph-Based"なアルゴリズムに基づくOntology、Clustering Methodが脚光を浴び始めた気がするけど、こうしたシーケンシングの可視化は、実効的にData-Drivenの代替手段と言えるのかな
□ Mathematical Model Resolves Decade-Old Debate On Regulation Of Protein Production By MicroRNAs In Cells
>> http://www.medicalnewstoday.com/releases/248486.php

□ genetics_blog:
Poster (PNG): "Visualization and ?#DataMining? of Integrated Whole-Genome Data using Self-Organizing Maps" ?#ISMB? ?#KN3?
□ Phil_at_OeRC:
?#ISMB? ?#KN3?: BW visualization of ENCODE data using SOM unwrapping doughnuts http://woldlab.caltech.edu/encodesom/
□ iGenomics:
Barbara Wold: RNA-Seq wet lab challenges - how to get full length transcripts: ave 2581bp, median 1592bp based GENCODE 17 ?#ismb? ?#KN3?
□ iGenomics:
Should BIoinformatics Cores have a "standard" pipelines? (I would argue to make them reproducible pipelines) ?#ismb? ?#WK?
□ figshare:
Raw ?#exome? files on ?#figshare? integrated into new article on @f1000research: http://shar.es/teAJz ?#openaccess? ?#opendata? ?#openscience?
ISMB見てても、今年度は次世代シーケンシング、pipelineはIntegrateしてもIntegrateしても。。integrateの為のVariousツールが出てくるよね。メーカー間の業務提携による解析ワークフローのプロトコル最適化もそう。収斂と拡散、どっちに傾向?
□ Sequencing low diversity libraries on Illumina MiSeq: 16S Seq: low yields, low per-base quality scores random libraries http://pathogenomics.bham.ac.uk/blog/2012/08/sequencing-low-diversity-libraries-on-illumina-miseq/ …
□ Xpression ? An intergrated RNA-seq pipeline
>> http://depts.washington.edu/cshlab/xpression/html/rnaseq.shtml
integrated solution for processing next-gen sequencing data derived from various types of samples, such as samples mapped to draft genomes and multiplexed samples.
□ MyMiner: a web application for computer-assisted biocuration and text annotation.
>> http://www.ncbi.nlm.nih.gov/m/pubmed/22789588/
MyMiner, a free and user-friendly text annotation tool aimed to assist in carrying out the main biocuration tasks and to provide labelled data for the development of text mining systems. MyMiner allows easy classification and labelling of textual data according to user-specified classes as well as predefined biological entities. The usefulness and efficiency of this application has been tested for a range of real-life annotation scenarios of various research topics.
□ Crystals, Information And The Origin of Life
>> http://t.co/Kxqa6lsj
The combination of information theory and crystallography should lead to a better understanding of DNA molecules, cells and perhaps even complex living things like humans
□ Beyond crystals: the dialectic of materials and information
>> http://rsta.royalsocietypublishing.org/content/370/1969/2807.full
the possibility of the artificial construction of a synthetic living system, different from biological life, but having many or all of the same properties. Interactions are essentially nonlinear and collective. Structures begin to have an evolutionary history with episodes of symbiosis.
Underlying all the structures are constraints of time and space. Through hierarchization, a more general principle than the periodicity of crystals, structures may be found within structures on different scales. We must integrate unifying concepts from dynamical systems and information th










