You've gotta learn, realize, and then die.
??学び 気づき 死ね

□ Constraints on the Universe as a Numerical Simulation
>> http://arxiv.org/pdf/1210.1847v2.pdf
□ InsideScience:
http://bit.ly/Z6uhzy - What if Reality Was Just Sim Universe? Physicists propose a test. #ISNS
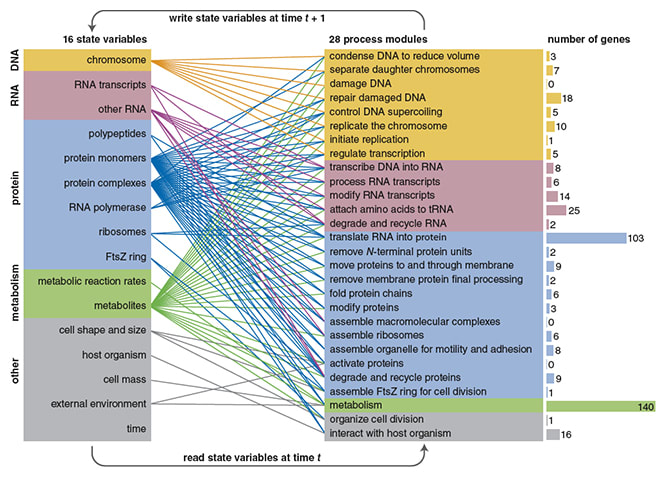
□ Imitation of Life: Can a computer program reproduce everything that happens inside a living cell?
>> http://www.americanscientist.org/issues/id.15983,y.2013,no.1,content.true,page.1,css.print/issue.aspx


□ NatureBiotech:
Analysis: A gene ontology inferred from molecular networks - Dutkowski et al.
>> http://bit.ly/12vNZCM
NeXO: The network-extracted ontology contains 4,123 biological terms and 5,766 term-term relations: 分子ネットワーク推量オントロジー

□ EMdeCODE: a novel algorithm capable of reading words of epigenetic code to predict enhancers & retroviral integration sites and to identify H3R2me1 as a distinctive mark of coding versus non-coding genes
>> http://m.nar.oxfordjournals.org/content/early/2012/12/11/nar.gks1214.full

□ GENOME-WIDE ASSOCIATION STUDIES ON HADOOP: A Cloud Epistasis cOmputing/FastTagger: An efficient algorithm for SNP
>> http://www.comp.nus.edu.sg/~wongls/psZ/wangyue-thesis.pdf
GENOME-WIDE ASSOCIATION STUDIES ON HADOOP: Hadoopを用いたゲノムワイド関連解析技術の蓄積。エピスタシス検出法、高次元特性のためのPaRFRアルゴリズム、RuleImputeによる計算精度向上
□ morungos:
Paper mentioned in personalized medicine session useful for many large scale hypothesis testing studies http://m.pnas.org/content/100/16/9440.full … #psb2013
□ Bioinformatic5:
Delivering the promise of public health genomics (thanks to @Karelman) http://bit.ly/100zwkA
□ BiomarkerCmns:
Two Major Reference Labs Sign With Critical Diagnostics http://tinyurl.com/9wvb9xn
□ Cost-effective GPU-Grid for Genome-wide Epistasis Calculations: Sequential epistatsis calculations > GPU using CUDA: http://www.ncbi.nlm.nih.gov/m/pubmed/23223640/

□ SemScape: Visualizating Semantic Web Data Landscapes with Cytoscape 3.0
>> http://figshare.com/articles/GSOC-Poster.pdf/103763

□ genefish:
[tumblrpic] cytoscape-publications: Comprehensive Analysis of Host Cellular Interactions with Human Papillomavir... pic.twitter.com/c1Q93HG1

□ DOAF: A Framework for Annotating Human Genome in Disease Context:
>> http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0049686
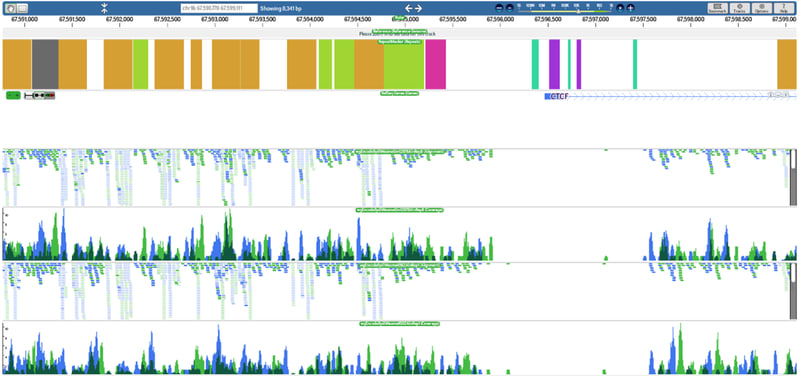
□ dnanexus:
Metapatterns used to explain impressive chromatin diversity on a regulatory level. Published in @genomeresearch http://ow.ly/fZnUV
the Clustered AGgregation Tool (CAGT): a new tool for pattern discovery from DNAnexus.


□ crAss: Reference-independent comparative metagenomics using cross-assembly:
>> http://bioinformatics.oxfordjournals.org/content/28/24/3225.full
□ HumanOrigins:
Most rare human DNA mutations arose in the last 5000 yrs. "We are freshly bursting with the raw material of evolution."
>> http://ow.ly/gezSz
・ Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants
>> http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11690.html
□ TrueSight: New algorithm for splice junction detection using RNA-seq: compare with TopHat, MapSplice & PASSion via Cufflinks
>> http://nar.oxfordjournals.org/content/early/2012/12/18/nar.gks1311.full
□ minerva and minepy: a C engine for the MINE suite and its R, Python and MATLAB wrappers
>> http://m.bioinformatics.oxfordjournals.org/content/early/2012/12/14/bioinformatics.bts707.short
□ druvus:
DNA cancer database plan prompts 'major concerns' http://zite.to/UlqQxY #sequencing
□ buildmodels:
3D Snapshots Reveal How Transcription Begins http://www.eurekalert.org/pub_releases/2012-10/ru-rru101512.php … PDB IDs 4g7o 4g7h 4g7z http://www.rcsb.org/pdb/explore/explore.do?structureId=4G7O … pic.twitter.com/X4tAAzlA
□ jocalynclark:
Global Burden of Disease estimates can become a global public good only if the data are made public http://bit.ly/UhwW2i #GBD2010

□ ISCB-Asia/SCCG 2012: (Dec17-19 Shenzen) #ISCBAsia
>> http://www.iscb.org/iscb-asia-sccg2012-conference-info
??学び 気づき 死ね

□ Constraints on the Universe as a Numerical Simulation
>> http://arxiv.org/pdf/1210.1847v2.pdf
Observable consequences of the hypothesis that the observed universe is a numerical simulation performed on a cubic space-time lattice or grid are explored. The simulation scenario is first motivated by extrapolating current trends in computational resource requirements for lattice QCD into the future. Using the historical development of lattice gauge theory technology as a guide, we assume that our universe is an early numerical simulation with unimproved Wilson fermion discretization and investigate potentially-observable consequences.この宇宙がシミュレートされたものであることをシミュレートする観測実験。立方時空格子における計算リソース要件とフェルミオン離散化
Among the observables that are considered are the muon g - 2 and the current differences between determinations of α, but the most stringent bound on the inverse lattice spacing of the universe, b-1>? 1011 GeV, is derived from the high-energy cut off of the cosmic ray spectrum. The numerical simulation scenario could reveal itself in the distributions of the highest energy cosmic rays exhibiting a degree of rotational symmetry breaking that reflects the structure of the underlying lattice.
□ InsideScience:
http://bit.ly/Z6uhzy - What if Reality Was Just Sim Universe? Physicists propose a test. #ISNS

□ Imitation of Life: Can a computer program reproduce everything that happens inside a living cell?
>> http://www.americanscientist.org/issues/id.15983,y.2013,no.1,content.true,page.1,css.print/issue.aspx
The WholeCell model is based on data collected from 900 publications. Some 1,900 numerical values were extracted from these sources to become parameters of the model. This is an impressive compendium, which anchors the simulation in real data.全細胞シミュレーション


□ NatureBiotech:
Analysis: A gene ontology inferred from molecular networks - Dutkowski et al.
>> http://bit.ly/12vNZCM
NeXO: The network-extracted ontology contains 4,123 biological terms and 5,766 term-term relations: 分子ネットワーク推量オントロジー
The network-extracted ontology (NeXO) contains 4,123 biological terms and 5,766 term-term relations, capturing 58% of known cellular components. We also explore robust NeXO terms and term relations that were initially not cataloged in GO, a number of which have now been added based on our analysis.
Using quantitative genetic interaction profiling and chemogenomics, we find further support for many of the uncharacterized terms identified by NeXO, including multisubunit structures related to protein trafficking or mitochondrial function. This work enables a shift from using ontologies to evaluate data to using data to construct and evaluate ontologies.

□ EMdeCODE: a novel algorithm capable of reading words of epigenetic code to predict enhancers & retroviral integration sites and to identify H3R2me1 as a distinctive mark of coding versus non-coding genes
>> http://m.nar.oxfordjournals.org/content/early/2012/12/11/nar.gks1214.full
EMdeCODE densities improve significantly the prediction of enhancer loci and retroviral integration sites with respect to previous methods. Importantly, it can also be used to extract distinctive factors between two arbitrary conditions. Indeed EMdeCODE identifies unexpected epigenetic profiles specific for coding versus non-coding RNA, pointing towards a new role for H3R2me1 in coding regions.

□ GENOME-WIDE ASSOCIATION STUDIES ON HADOOP: A Cloud Epistasis cOmputing/FastTagger: An efficient algorithm for SNP
>> http://www.comp.nus.edu.sg/~wongls/psZ/wangyue-thesis.pdf
This thesis explores the data analysis involved in genome-wide association stud- ies (GWAS) using Hadoop technologies and data mining techniques. GWAS is amongst the most popular study designs to identify potential genetic variants that are linked to the etiologies of diseases. In the future, GWAS will also play an important role in personalized medicine. The complex data analysis in GWAS calls for new technologies and techniques.
GENOME-WIDE ASSOCIATION STUDIES ON HADOOP: Hadoopを用いたゲノムワイド関連解析技術の蓄積。エピスタシス検出法、高次元特性のためのPaRFRアルゴリズム、RuleImputeによる計算精度向上
□ morungos:
Paper mentioned in personalized medicine session useful for many large scale hypothesis testing studies http://m.pnas.org/content/100/16/9440.full … #psb2013
□ Bioinformatic5:
Delivering the promise of public health genomics (thanks to @Karelman) http://bit.ly/100zwkA
□ BiomarkerCmns:
Two Major Reference Labs Sign With Critical Diagnostics http://tinyurl.com/9wvb9xn
□ Cost-effective GPU-Grid for Genome-wide Epistasis Calculations: Sequential epistatsis calculations > GPU using CUDA: http://www.ncbi.nlm.nih.gov/m/pubmed/23223640/

□ SemScape: Visualizating Semantic Web Data Landscapes with Cytoscape 3.0
>> http://figshare.com/articles/GSOC-Poster.pdf/103763
SemScape is developed as plugin for the latest version of Cytoscape, 3.0, which is released in beta at the time of this writing and presents a completely new architecture from previous versions. SemScape offers a range of functionalities, which are derived from what was implemented in RDFScape [2] and AGUIA [3]. However this system provides a newer, more eff cient and up to date implementation.

□ genefish:
[tumblrpic] cytoscape-publications: Comprehensive Analysis of Host Cellular Interactions with Human Papillomavir... pic.twitter.com/c1Q93HG1

□ DOAF: A Framework for Annotating Human Genome in Disease Context:
>> http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0049686
The Disease Ontology Annotation Framework (DOAF) to provide a comprehensive annotation of the human genome using the computable Disease Ontology (DO), the NCBO Annotator service and NCBI Gene Reference Into Function (GeneRIF). DOAF can keep the resulting knowledgebase current by periodically executing automatic pipeline to re-annotate the human genome using the latest DO and GeneRIF releases at any frequency such as daily or monthly.
DOAF provides a computable and programmable environment which enables large-scale and integrative analysis by working with external analytic software or online service platforms. A user-friendly web interface (doa.nubic.northwestern.edu) is implemented to allow users to efficiently query, download, and view disease annotations and the underlying evidences.

□ dnanexus:
Metapatterns used to explain impressive chromatin diversity on a regulatory level. Published in @genomeresearch http://ow.ly/fZnUV
the Clustered AGgregation Tool (CAGT): a new tool for pattern discovery from DNAnexus.
To improve accuracy, the team sequenced extremely deeply, ultimately generating some 5 billion reads on the SOLiD sequencing platform. “The data sets were incredibly massive,” Kundaje says. “Processing these data sets locally was quite a challenge.” The group turned to DNAnexus, uploading their sequence files to the cloud and preprocessing the data with the company’s probabilistic mapping tool. “DNAnexus made that process incredibly simple,"SOLiDで5億リードを生成、新手法で確率的マッピング


□ crAss: Reference-independent comparative metagenomics using cross-assembly:
>> http://bioinformatics.oxfordjournals.org/content/28/24/3225.full
crAss, a novel more direct approach based on the same concept, i.e. using cross-assembly of reads from different metagenomes to assess the degree of similarity between the sampled communities. Thus, cross-metagenome assembly enables a sensitive comparison of entities that are shared between samples, including viruses with no homology to known sequences.未知の配列によるキメラ・シーケンスの影響を制限する非リファレンス依存のメタゲノム解析
□ HumanOrigins:
Most rare human DNA mutations arose in the last 5000 yrs. "We are freshly bursting with the raw material of evolution."
>> http://ow.ly/gezSz
・ Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants
>> http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11690.html
approximately 73% of all protein-coding SNVs and approximately 86% of SNVs predicted to be deleterious arose in the past 5,000?10,000?years. The average age of deleterious SNVs varied significantly across molecular pathways, and disease genes contained a significantly higher proportion of recently arisen deleterious SNVs than other genes.人類の遺伝的変異は一万年で急速にポテンシャルを蓄積している
□ TrueSight: New algorithm for splice junction detection using RNA-seq: compare with TopHat, MapSplice & PASSion via Cufflinks
>> http://nar.oxfordjournals.org/content/early/2012/12/18/nar.gks1311.full
TrueSight, which for the first time combines RNA-seq read mapping quality and coding potential of genomic sequences into a unified model. The model is further utilized in a machine-learning approach to precisely identify SJs. Both simulations and real data evaluations showed that TrueSight achieved higher sensitivity and specificity than other methods. We applied TrueSight to new high coverage honey bee RNA-seq data to discover novel splice forms.
□ minerva and minepy: a C engine for the MINE suite and its R, Python and MATLAB wrappers
>> http://m.bioinformatics.oxfordjournals.org/content/early/2012/12/14/bioinformatics.bts707.short
a novel implementation in ANSI C of the MINE family of algorithms for computing maximal information-based measures of dependence between two variables in large datasets, with the aim of a low memory footprint and ease of integration within bioinformatics pipelines.
We provide the libraries minerva (with the R interface) and minepy for Python, MATLAB, Octave and C++. The C solution reduces the large memory requirement of the original Java implementation, has good upscaling properties, and offers a native parallelization for the R interface. Low memory requirements are demonstrated on the MINE benchmarks as well as on large (n=1340) microarray and Illumina GAII RNA-seq transcriptomics datasets.
□ druvus:
DNA cancer database plan prompts 'major concerns' http://zite.to/UlqQxY #sequencing
□ buildmodels:
3D Snapshots Reveal How Transcription Begins http://www.eurekalert.org/pub_releases/2012-10/ru-rru101512.php … PDB IDs 4g7o 4g7h 4g7z http://www.rcsb.org/pdb/explore/explore.do?structureId=4G7O … pic.twitter.com/X4tAAzlA
□ jocalynclark:
Global Burden of Disease estimates can become a global public good only if the data are made public http://bit.ly/UhwW2i #GBD2010

□ ISCB-Asia/SCCG 2012: (Dec17-19 Shenzen) #ISCBAsia
>> http://www.iscb.org/iscb-asia-sccg2012-conference-info
ISCB: computational tools for NGS, cancer informatics, and cloud and work flows for reproducible biology. ISCB is known for hosting a variety of international meetings including ISMB, while BGI of Shenzhen is one of the world's largest sequencing centers.
SCCG: The local and international scientific community is highly encouraged to participate in this extraordinary event that is bringing together some of the world’s thought leaders in the fields of computational biology, biomedical informatics, and genomics, as well as providing a forum for the presentation of the excellent research being d










