Daniel Hanisch, Alexander Zien, Ralf Zimmer and Thomas Lengauer
Co-clustering of biological networks and gene expression data
Bioinformatics Vol. 18 no. 90001 2002, Pages S145-S154
[PDF][Web Site]
・遺伝子ネットワーク推定法について。従来法は遺伝子発現量情報のみに基いていたのに対して、さらにBiological networksの情報を付加した方法を提案する。
・データ:Yeast, 時間点7点 [DeRisi,1997] Metabolic networkに着目。
・クラスタリング法:Hierarchical average linkage clustering [Eisen,1998]。閾値の決定はSilhouette-coefficient [Rousseeuw,1987]による。
・問題点「One major problem is that the boundaries of the resulting clusters are arbitrary to some degree. 」
・方法「In this paper, we propose a novel method that utilizes information in the form of biological networks in an integrated manner to improve the result of the clustering.」
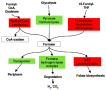
・「Biological networks relate genes, gene products or groups of those (e.g., protein complexes or protein families) to each other in the form of a graph (Bhalla and Iyengar,1999).」
・方法「Whereas conventional cluster methods rely on gene expression data alone, we propose combining measures derived from gene expression data and metrics on biological networks into a single distance function.」
・「The most popular choice in the case of time-series data is the Pearson correlation coefficient, as suggested by Eisen et al.(1998).」
Co-clustering of biological networks and gene expression data
Bioinformatics Vol. 18 no. 90001 2002, Pages S145-S154
[PDF][Web Site]
・遺伝子ネットワーク推定法について。従来法は遺伝子発現量情報のみに基いていたのに対して、さらにBiological networksの情報を付加した方法を提案する。
・データ:Yeast, 時間点7点 [DeRisi,1997] Metabolic networkに着目。
・クラスタリング法:Hierarchical average linkage clustering [Eisen,1998]。閾値の決定はSilhouette-coefficient [Rousseeuw,1987]による。
・問題点「One major problem is that the boundaries of the resulting clusters are arbitrary to some degree. 」
・方法「In this paper, we propose a novel method that utilizes information in the form of biological networks in an integrated manner to improve the result of the clustering.」
・「Biological networks relate genes, gene products or groups of those (e.g., protein complexes or protein families) to each other in the form of a graph (Bhalla and Iyengar,1999).」
・方法「Whereas conventional cluster methods rely on gene expression data alone, we propose combining measures derived from gene expression data and metrics on biological networks into a single distance function.」
・「The most popular choice in the case of time-series data is the Pearson correlation coefficient, as suggested by Eisen et al.(1998).」