#==========
Julia の修行をするときに,いろいろなプログラムを書き換えるのは有効な方法だ。
以下のプログラムを Julia に翻訳してみる。
数量化 I 類
http://aoki2.si.gunma-u.ac.jp/R/qt1.html
ファイル名: qt1.jl 関数名: qt1, printqt1, plotqt1
翻訳するときに書いたメモ
==========#
using Statistics, StatsBase, Rmath, LinearAlgebra, NamedArrays, Plots
function qt1(dat, y; vnames=[], vnamey="")
dat = hcat(dat, y)
nc, p = size(dat)
ncat = p - 1
length(vnames) > 0 || (vnames = ["x$i" for i = 1:ncat])
vnamey != "" || (vnamey = "dep. var.")
mx = [maximum(dat[:, i]) for i = 1:ncat]
start = vcat(0, cumsum(mx)[1:ncat - 1])
nobe = sum(mx)
x = zeros(nc, nobe - ncat + 1)
for i = 1:nc
x0 = []
for j = 1:ncat
zeros0 = zeros(mx[j])
zeros0[dat[i, j]] = 1
append!(x0, zeros0[2:end])
end
x[i, :] = vcat(x0, dat[i, end])
end
a = cov(x)
ndim = nobe - ncat
B = a[1:ndim, 1:ndim] \ a[ndim + 1, 1:ndim]
m = mean(x, dims=1)
constant = m[ndim + 1] - (m[1:ndim]' * B)
predicted = x[:, 1:ndim] * B .+ constant
observed = x[:, ndim + 1]
residuals = observed .- predicted
s = sum(x, dims=1)
en = 0
coef = []
coefnames = []
for i = 1:ncat
st = en + 1
en = st + mx[i] - 2
target = st:en
tempmean = (s[target]' * B[target]) / nc
constant = constant + tempmean
append!(coef, -tempmean, B[target] .- tempmean)
append!(coefnames, ["$(vnames[i]).$j" for j = 1:mx[i]])
end
append!(coef, constant)
append!(coefnames, ["constant"])
q = ["a", "b"]
par = zeros(nc, ncat)
for j in 1:nc
en = 0
for i in 1:ncat
st = en + 1
en = st + mx[i] - 2
target = st:en
par[j, i] = x[j, target]' * B[target]
end
end
par = hcat(par, observed)
r = cor(par)
i = inv(r)
d = diag(i)
partialcor = (-i ./ sqrt.(d * d'))[ncat + 1, 1:ncat]
partialt = abs.(partialcor) .* sqrt.((nc - ncat - 1) ./ (1 .- partialcor .^ 2))
partialp = pt.(partialt, nc - ncat - 1, false) .* 2
Dict(:coef => coef, :coefnames => coefnames,
:r => r, :partialcor => partialcor, :partialt => partialt,
:partialp => partialp, :predicted => predicted,
:observed => observed, :residuals => residuals,
:vnames => vnames, :vnamey => vnamey)
end
function printqt1(obj)
println("\ncategory score\n", NamedArray(reshape(obj[:coef], :, 1),
(obj[:coefnames], ["category score"])))
names = vcat(obj[:vnames], obj[:vnamey])
println("\ncorrelation coefficient\n", NamedArray(obj[:r], (names, names)))
println("\npartial correlation coeficiennt\n",
NamedArray(hcat(obj[:partialcor], obj[:partialt], obj[:partialp]),
(obj[:vnames], ["partial corr.", "t value", "p value"])))
println("\nprediction\n",
NamedArray(hcat(obj[:observed], obj[:predicted], obj[:residuals]),
(1:length(obj[:observed]), ["observed", "predicted", "residuals"])))
end
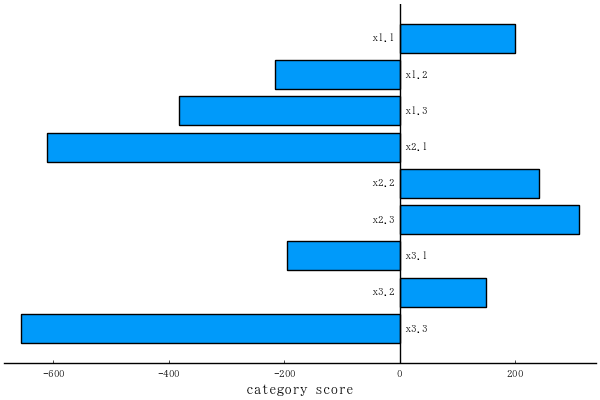
function plotqt1(obj; which = "categoryscore") # or "fitness"
pyplot()
if which == "categoryscore"
coefficients = obj[:coef][end-1:-1:1]
plt = bar(coefficients, orientation=:h, grid=false,
yshowaxis=false, yticks=false,
xlabel="category score", label="")
cname = [" $s " for s in obj[:coefnames][end-1:-1:1]]
align = [c > 0 ? :right : :left for c in coefficients]
annotate!.(0, 1:length(cname), text.(cname, align, 8))
vline!([0], color=:black, label="")
else
minx, maxx = extrema(vcat(obj[:predicted], obj[:observed]))
w = 0.05(maxx - minx)
minx -= w
maxx += w
plt = scatter(obj[:predicted], obj[:observed], grid=false,
tick_direction=:out, xlabel = "predicted",
ylabel = "observed", aspect_ratio = 1,
lims=(minx, maxx), size=(600, 600), label="")
plot!([minx, maxx], [minx, maxx], label="")
end
display(plt)
end
dat = [ 1 2 2
3 2 2
1 2 2
1 1 1
2 3 2
1 3 3
2 2 2
1 1 1
3 2 2
1 1 2];
y = [6837, 7397, 7195, 6710, 6670, 6279, 6601, 4929, 5471, 6164];
obj = qt1(dat, y)
printqt1(obj)
#=====
category score
10×1 Named Matrix{Any}
A ╲ B │ category score
─────────┼───────────────
x1.1 │ 199.4
x1.2 │ -215.6
x1.3 │ -382.6
x2.1 │ -610.2
x2.2 │ 241.8
x2.3 │ 310.8
x3.1 │ -195.0
x3.2 │ 149.5
x3.3 │ -656.5
constant │ 6425.3
correlation coefficient
4×4 Named Matrix{Float64}
A ╲ B │ x1 x2 x3 dep. var.
──────────┼───────────────────────────────────────────
x1 │ 1.0 -0.510664 -0.463184 -0.103277
x2 │ -0.510664 1.0 0.164788 0.440587
x3 │ -0.463184 0.164788 1.0 0.290518
dep. var. │ -0.103277 0.440587 0.290518 1.0
partial correlation coeficiennt
3×3 Named Matrix{Float64}
A ╲ B │ partial corr. t value p value
──────┼────────────────────────────────────────────
x1 │ 0.308748 0.795121 0.456835
x2 │ 0.500942 1.41777 0.206039
x3 │ 0.358395 0.940354 0.383335
prediction
10×3 Named Matrix{Float64}
A ╲ B │ observed predicted residuals
──────┼─────────────────────────────────────────
1 │ 6837.0 7016.0 -179.0
2 │ 7397.0 6434.0 963.0
3 │ 7195.0 7016.0 179.0
4 │ 6710.0 5819.5 890.5
5 │ 6670.0 6670.0 0.0
6 │ 6279.0 6279.0 0.0
7 │ 6601.0 6601.0 -9.09495e-13
8 │ 4929.0 5819.5 -890.5
9 │ 5471.0 6434.0 -963.0
10 │ 6164.0 6164.0 -9.09495e-13
=====#
plotqt1(obj) # savefig("fig1.png")
plotqt1(obj, which="fitness") # savefig("fig2.png")